Merge remote-tracking branch 'origin/main' into fix-ollama-integration
This commit is contained in:
127
.gitignore
vendored
127
.gitignore
vendored
@@ -1,121 +1,8 @@
|
||||
# Byte-compiled / optimized / DLL files
|
||||
__pycache__/
|
||||
*.py[cod]
|
||||
*$py.class
|
||||
|
||||
# C extensions
|
||||
*.so
|
||||
|
||||
# Distribution / packaging
|
||||
.Python
|
||||
env/
|
||||
venv/
|
||||
ENV/
|
||||
env.bak/
|
||||
venv.bak/
|
||||
*.egg
|
||||
*.egg-info/
|
||||
dist/
|
||||
build/
|
||||
*.whl
|
||||
|
||||
# PyInstaller
|
||||
# Usually these files are written by a python script from a template
|
||||
# before PyInstaller builds the exe, so as to inject date/other infos into it.
|
||||
*.manifest
|
||||
*.spec
|
||||
|
||||
# Installer logs
|
||||
pip-log.txt
|
||||
pip-delete-this-directory.txt
|
||||
|
||||
# Unit test / coverage reports
|
||||
htmlcov/
|
||||
.tox/
|
||||
.nox/
|
||||
.coverage
|
||||
.coverage.*
|
||||
.cache
|
||||
nosetests.xml
|
||||
coverage.xml
|
||||
*.cover
|
||||
*.py,cover
|
||||
.hypothesis/
|
||||
|
||||
# Translations
|
||||
*.mo
|
||||
*.pot
|
||||
|
||||
# Django stuff:
|
||||
*.log
|
||||
local_settings.py
|
||||
db.sqlite3
|
||||
db.sqlite3-journal
|
||||
|
||||
# Flask stuff:
|
||||
instance/
|
||||
.webassets-cache
|
||||
|
||||
# Scrapy stuff:
|
||||
.scrapy
|
||||
|
||||
# Sphinx documentation
|
||||
docs/_build/
|
||||
|
||||
# PyBuilder
|
||||
target/
|
||||
|
||||
# Jupyter Notebook
|
||||
.ipynb_checkpoints
|
||||
|
||||
# IPython
|
||||
profile_default/
|
||||
ipython_config.py
|
||||
|
||||
# pyenv
|
||||
.python-version
|
||||
|
||||
# celery beat schedule file
|
||||
celerybeat-schedule
|
||||
|
||||
# SageMath parsed files
|
||||
*.sage.py
|
||||
|
||||
# Environments
|
||||
.env
|
||||
.env.*
|
||||
.venv
|
||||
.venv.*
|
||||
env/
|
||||
venv/
|
||||
ENV/
|
||||
env.bak/
|
||||
venv.bak/
|
||||
|
||||
# Spyder project settings
|
||||
.spyderproject
|
||||
.spyderworkspace
|
||||
|
||||
# Rope project settings
|
||||
.ropeproject
|
||||
|
||||
# mkdocs documentation
|
||||
/site
|
||||
|
||||
# mypy
|
||||
.mypy_cache/
|
||||
.dmypy.json
|
||||
dmypy.json
|
||||
|
||||
# Pyre type checker
|
||||
.pyre/
|
||||
|
||||
# pytype static type analyzer
|
||||
.pytype/
|
||||
|
||||
# Cython debug symbols
|
||||
cython_debug/
|
||||
|
||||
# Example files
|
||||
book.txt
|
||||
__pycache__
|
||||
*.egg-info
|
||||
dickens/
|
||||
book.txt
|
||||
lightrag-dev/
|
||||
.idea/
|
||||
dist/
|
||||
.venv/
|
||||
|
||||
22
.pre-commit-config.yaml
Normal file
22
.pre-commit-config.yaml
Normal file
@@ -0,0 +1,22 @@
|
||||
repos:
|
||||
- repo: https://github.com/pre-commit/pre-commit-hooks
|
||||
rev: v5.0.0
|
||||
hooks:
|
||||
- id: trailing-whitespace
|
||||
- id: end-of-file-fixer
|
||||
- id: requirements-txt-fixer
|
||||
|
||||
|
||||
- repo: https://github.com/astral-sh/ruff-pre-commit
|
||||
rev: v0.6.4
|
||||
hooks:
|
||||
- id: ruff-format
|
||||
- id: ruff
|
||||
args: [--fix]
|
||||
|
||||
|
||||
- repo: https://github.com/mgedmin/check-manifest
|
||||
rev: "0.49"
|
||||
hooks:
|
||||
- id: check-manifest
|
||||
stages: [manual]
|
||||
252
README.md
252
README.md
@@ -16,16 +16,17 @@
|
||||
<a href="https://pypi.org/project/lightrag-hku/"><img src="https://img.shields.io/pypi/v/lightrag-hku.svg"></a>
|
||||
<a href="https://pepy.tech/project/lightrag-hku"><img src="https://static.pepy.tech/badge/lightrag-hku/month"></a>
|
||||
</p>
|
||||
|
||||
|
||||
This repository hosts the code of LightRAG. The structure of this code is based on [nano-graphrag](https://github.com/gusye1234/nano-graphrag).
|
||||
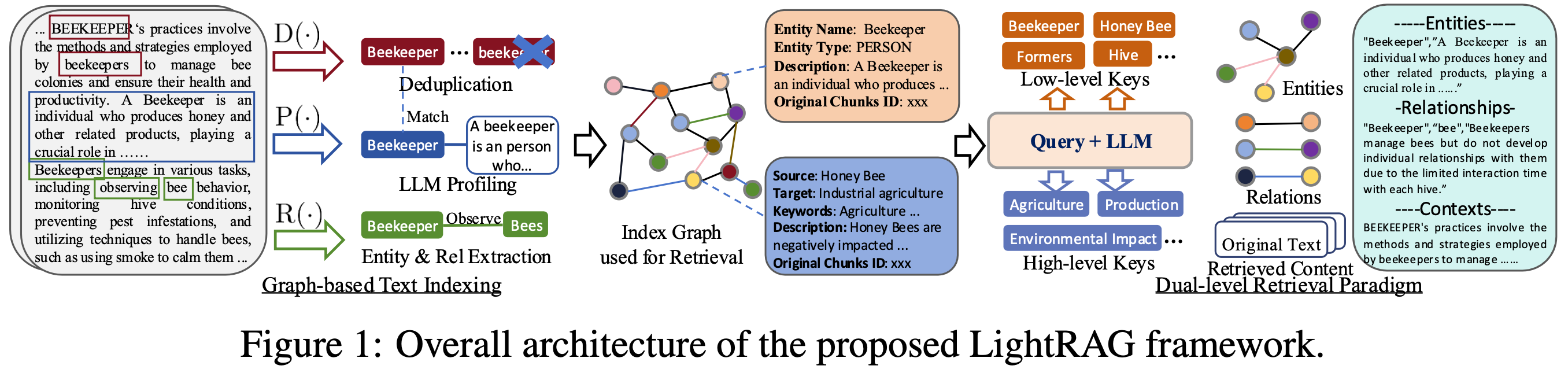
|
||||
</div>
|
||||
|
||||
## 🎉 News
|
||||
## 🎉 News
|
||||
- [x] [2024.10.20]🎯🎯📢📢We’ve added a new feature to LightRAG: Graph Visualization.
|
||||
- [x] [2024.10.18]🎯🎯📢📢We’ve added a link to a [LightRAG Introduction Video](https://youtu.be/oageL-1I0GE). Thanks to the author!
|
||||
- [x] [2024.10.17]🎯🎯📢📢We have created a [Discord channel](https://discord.gg/mvsfu2Tg)! Welcome to join for sharing and discussions! 🎉🎉
|
||||
- [x] [2024.10.16]🎯🎯📢📢LightRAG now supports [Ollama models](https://github.com/HKUDS/LightRAG?tab=readme-ov-file#quick-start)!
|
||||
- [x] [2024.10.15]🎯🎯📢📢LightRAG now supports [Hugging Face models](https://github.com/HKUDS/LightRAG?tab=readme-ov-file#quick-start)!
|
||||
- [x] [2024.10.16]🎯🎯📢📢LightRAG now supports [Ollama models](https://github.com/HKUDS/LightRAG?tab=readme-ov-file#quick-start)!
|
||||
- [x] [2024.10.15]🎯🎯📢📢LightRAG now supports [Hugging Face models](https://github.com/HKUDS/LightRAG?tab=readme-ov-file#quick-start)!
|
||||
|
||||
## Install
|
||||
|
||||
@@ -47,12 +48,21 @@ pip install lightrag-hku
|
||||
```bash
|
||||
curl https://raw.githubusercontent.com/gusye1234/nano-graphrag/main/tests/mock_data.txt > ./book.txt
|
||||
```
|
||||
Use the below Python snippet to initialize LightRAG and perform queries:
|
||||
Use the below Python snippet (in a script) to initialize LightRAG and perform queries:
|
||||
|
||||
```python
|
||||
from lightrag import LightRAG, QueryParam
|
||||
from lightrag.llm import gpt_4o_mini_complete, gpt_4o_complete
|
||||
|
||||
#########
|
||||
# Uncomment the below two lines if running in a jupyter notebook to handle the async nature of rag.insert()
|
||||
# import nest_asyncio
|
||||
# nest_asyncio.apply()
|
||||
#########
|
||||
|
||||
WORKING_DIR = "./dickens"
|
||||
|
||||
|
||||
WORKING_DIR = "./dickens"
|
||||
|
||||
if not os.path.exists(WORKING_DIR):
|
||||
@@ -83,7 +93,7 @@ print(rag.query("What are the top themes in this story?", param=QueryParam(mode=
|
||||
<details>
|
||||
<summary> Using Open AI-like APIs </summary>
|
||||
|
||||
LightRAG also support Open AI-like chat/embeddings APIs:
|
||||
* LightRAG also supports Open AI-like chat/embeddings APIs:
|
||||
```python
|
||||
async def llm_model_func(
|
||||
prompt, system_prompt=None, history_messages=[], **kwargs
|
||||
@@ -120,8 +130,8 @@ rag = LightRAG(
|
||||
|
||||
<details>
|
||||
<summary> Using Hugging Face Models </summary>
|
||||
|
||||
If you want to use Hugging Face models, you only need to set LightRAG as follows:
|
||||
|
||||
* If you want to use Hugging Face models, you only need to set LightRAG as follows:
|
||||
```python
|
||||
from lightrag.llm import hf_model_complete, hf_embedding
|
||||
from transformers import AutoModel, AutoTokenizer
|
||||
@@ -136,7 +146,7 @@ rag = LightRAG(
|
||||
embedding_dim=384,
|
||||
max_token_size=5000,
|
||||
func=lambda texts: hf_embedding(
|
||||
texts,
|
||||
texts,
|
||||
tokenizer=AutoTokenizer.from_pretrained("sentence-transformers/all-MiniLM-L6-v2"),
|
||||
embed_model=AutoModel.from_pretrained("sentence-transformers/all-MiniLM-L6-v2")
|
||||
)
|
||||
@@ -147,8 +157,9 @@ rag = LightRAG(
|
||||
|
||||
<details>
|
||||
<summary> Using Ollama Models </summary>
|
||||
If you want to use Ollama models, you only need to set LightRAG as follows:
|
||||
|
||||
* If you want to use Ollama models, you only need to set LightRAG as follows:
|
||||
|
||||
```python
|
||||
from lightrag.llm import ollama_model_complete, ollama_embedding
|
||||
|
||||
@@ -162,12 +173,35 @@ rag = LightRAG(
|
||||
embedding_dim=768,
|
||||
max_token_size=8192,
|
||||
func=lambda texts: ollama_embedding(
|
||||
texts,
|
||||
texts,
|
||||
embed_model="nomic-embed-text"
|
||||
)
|
||||
),
|
||||
)
|
||||
```
|
||||
|
||||
* Increasing the `num_ctx` parameter:
|
||||
|
||||
1. Pull the model:
|
||||
```python
|
||||
ollama pull qwen2
|
||||
```
|
||||
|
||||
2. Display the model file:
|
||||
```python
|
||||
ollama show --modelfile qwen2 > Modelfile
|
||||
```
|
||||
|
||||
3. Edit the Modelfile by adding the following line:
|
||||
```python
|
||||
PARAMETER num_ctx 32768
|
||||
```
|
||||
|
||||
4. Create the modified model:
|
||||
```python
|
||||
ollama create -f Modelfile qwen2m
|
||||
```
|
||||
|
||||
</details>
|
||||
|
||||
### Batch Insert
|
||||
@@ -185,16 +219,171 @@ rag = LightRAG(working_dir="./dickens")
|
||||
with open("./newText.txt") as f:
|
||||
rag.insert(f.read())
|
||||
```
|
||||
|
||||
### Graph Visualization
|
||||
|
||||
<details>
|
||||
<summary> Graph visualization with html </summary>
|
||||
|
||||
* The following code can be found in `examples/graph_visual_with_html.py`
|
||||
|
||||
```python
|
||||
import networkx as nx
|
||||
from pyvis.network import Network
|
||||
|
||||
# Load the GraphML file
|
||||
G = nx.read_graphml('./dickens/graph_chunk_entity_relation.graphml')
|
||||
|
||||
# Create a Pyvis network
|
||||
net = Network(notebook=True)
|
||||
|
||||
# Convert NetworkX graph to Pyvis network
|
||||
net.from_nx(G)
|
||||
|
||||
# Save and display the network
|
||||
net.show('knowledge_graph.html')
|
||||
```
|
||||
|
||||
</details>
|
||||
|
||||
<details>
|
||||
<summary> Graph visualization with Neo4j </summary>
|
||||
|
||||
* The following code can be found in `examples/graph_visual_with_neo4j.py`
|
||||
|
||||
```python
|
||||
import os
|
||||
import json
|
||||
from lightrag.utils import xml_to_json
|
||||
from neo4j import GraphDatabase
|
||||
|
||||
# Constants
|
||||
WORKING_DIR = "./dickens"
|
||||
BATCH_SIZE_NODES = 500
|
||||
BATCH_SIZE_EDGES = 100
|
||||
|
||||
# Neo4j connection credentials
|
||||
NEO4J_URI = "bolt://localhost:7687"
|
||||
NEO4J_USERNAME = "neo4j"
|
||||
NEO4J_PASSWORD = "your_password"
|
||||
|
||||
def convert_xml_to_json(xml_path, output_path):
|
||||
"""Converts XML file to JSON and saves the output."""
|
||||
if not os.path.exists(xml_path):
|
||||
print(f"Error: File not found - {xml_path}")
|
||||
return None
|
||||
|
||||
json_data = xml_to_json(xml_path)
|
||||
if json_data:
|
||||
with open(output_path, 'w', encoding='utf-8') as f:
|
||||
json.dump(json_data, f, ensure_ascii=False, indent=2)
|
||||
print(f"JSON file created: {output_path}")
|
||||
return json_data
|
||||
else:
|
||||
print("Failed to create JSON data")
|
||||
return None
|
||||
|
||||
def process_in_batches(tx, query, data, batch_size):
|
||||
"""Process data in batches and execute the given query."""
|
||||
for i in range(0, len(data), batch_size):
|
||||
batch = data[i:i + batch_size]
|
||||
tx.run(query, {"nodes": batch} if "nodes" in query else {"edges": batch})
|
||||
|
||||
def main():
|
||||
# Paths
|
||||
xml_file = os.path.join(WORKING_DIR, 'graph_chunk_entity_relation.graphml')
|
||||
json_file = os.path.join(WORKING_DIR, 'graph_data.json')
|
||||
|
||||
# Convert XML to JSON
|
||||
json_data = convert_xml_to_json(xml_file, json_file)
|
||||
if json_data is None:
|
||||
return
|
||||
|
||||
# Load nodes and edges
|
||||
nodes = json_data.get('nodes', [])
|
||||
edges = json_data.get('edges', [])
|
||||
|
||||
# Neo4j queries
|
||||
create_nodes_query = """
|
||||
UNWIND $nodes AS node
|
||||
MERGE (e:Entity {id: node.id})
|
||||
SET e.entity_type = node.entity_type,
|
||||
e.description = node.description,
|
||||
e.source_id = node.source_id,
|
||||
e.displayName = node.id
|
||||
REMOVE e:Entity
|
||||
WITH e, node
|
||||
CALL apoc.create.addLabels(e, [node.entity_type]) YIELD node AS labeledNode
|
||||
RETURN count(*)
|
||||
"""
|
||||
|
||||
create_edges_query = """
|
||||
UNWIND $edges AS edge
|
||||
MATCH (source {id: edge.source})
|
||||
MATCH (target {id: edge.target})
|
||||
WITH source, target, edge,
|
||||
CASE
|
||||
WHEN edge.keywords CONTAINS 'lead' THEN 'lead'
|
||||
WHEN edge.keywords CONTAINS 'participate' THEN 'participate'
|
||||
WHEN edge.keywords CONTAINS 'uses' THEN 'uses'
|
||||
WHEN edge.keywords CONTAINS 'located' THEN 'located'
|
||||
WHEN edge.keywords CONTAINS 'occurs' THEN 'occurs'
|
||||
ELSE REPLACE(SPLIT(edge.keywords, ',')[0], '\"', '')
|
||||
END AS relType
|
||||
CALL apoc.create.relationship(source, relType, {
|
||||
weight: edge.weight,
|
||||
description: edge.description,
|
||||
keywords: edge.keywords,
|
||||
source_id: edge.source_id
|
||||
}, target) YIELD rel
|
||||
RETURN count(*)
|
||||
"""
|
||||
|
||||
set_displayname_and_labels_query = """
|
||||
MATCH (n)
|
||||
SET n.displayName = n.id
|
||||
WITH n
|
||||
CALL apoc.create.setLabels(n, [n.entity_type]) YIELD node
|
||||
RETURN count(*)
|
||||
"""
|
||||
|
||||
# Create a Neo4j driver
|
||||
driver = GraphDatabase.driver(NEO4J_URI, auth=(NEO4J_USERNAME, NEO4J_PASSWORD))
|
||||
|
||||
try:
|
||||
# Execute queries in batches
|
||||
with driver.session() as session:
|
||||
# Insert nodes in batches
|
||||
session.execute_write(process_in_batches, create_nodes_query, nodes, BATCH_SIZE_NODES)
|
||||
|
||||
# Insert edges in batches
|
||||
session.execute_write(process_in_batches, create_edges_query, edges, BATCH_SIZE_EDGES)
|
||||
|
||||
# Set displayName and labels
|
||||
session.run(set_displayname_and_labels_query)
|
||||
|
||||
except Exception as e:
|
||||
print(f"Error occurred: {e}")
|
||||
|
||||
finally:
|
||||
driver.close()
|
||||
|
||||
if __name__ == "__main__":
|
||||
main()
|
||||
```
|
||||
|
||||
</details>
|
||||
|
||||
## Evaluation
|
||||
### Dataset
|
||||
The dataset used in LightRAG can be download from [TommyChien/UltraDomain](https://huggingface.co/datasets/TommyChien/UltraDomain).
|
||||
The dataset used in LightRAG can be downloaded from [TommyChien/UltraDomain](https://huggingface.co/datasets/TommyChien/UltraDomain).
|
||||
|
||||
### Generate Query
|
||||
LightRAG uses the following prompt to generate high-level queries, with the corresponding code located in `example/generate_query.py`.
|
||||
LightRAG uses the following prompt to generate high-level queries, with the corresponding code in `example/generate_query.py`.
|
||||
|
||||
<details>
|
||||
<summary> Prompt </summary>
|
||||
|
||||
|
||||
```python
|
||||
Given the following description of a dataset:
|
||||
|
||||
@@ -219,18 +408,18 @@ Output the results in the following structure:
|
||||
...
|
||||
```
|
||||
</details>
|
||||
|
||||
|
||||
### Batch Eval
|
||||
To evaluate the performance of two RAG systems on high-level queries, LightRAG uses the following prompt, with the specific code available in `example/batch_eval.py`.
|
||||
|
||||
<details>
|
||||
<summary> Prompt </summary>
|
||||
|
||||
|
||||
```python
|
||||
---Role---
|
||||
You are an expert tasked with evaluating two answers to the same question based on three criteria: **Comprehensiveness**, **Diversity**, and **Empowerment**.
|
||||
---Goal---
|
||||
You will evaluate two answers to the same question based on three criteria: **Comprehensiveness**, **Diversity**, and **Empowerment**.
|
||||
You will evaluate two answers to the same question based on three criteria: **Comprehensiveness**, **Diversity**, and **Empowerment**.
|
||||
|
||||
- **Comprehensiveness**: How much detail does the answer provide to cover all aspects and details of the question?
|
||||
- **Diversity**: How varied and rich is the answer in providing different perspectives and insights on the question?
|
||||
@@ -294,7 +483,7 @@ Output your evaluation in the following JSON format:
|
||||
| **Empowerment** | 36.69% | **63.31%** | 45.09% | **54.91%** | 42.81% | **57.19%** | **52.94%** | 47.06% |
|
||||
| **Overall** | 43.62% | **56.38%** | 45.98% | **54.02%** | 45.70% | **54.30%** | **51.86%** | 48.14% |
|
||||
|
||||
## Reproduce
|
||||
## Reproduce
|
||||
All the code can be found in the `./reproduce` directory.
|
||||
|
||||
### Step-0 Extract Unique Contexts
|
||||
@@ -302,7 +491,7 @@ First, we need to extract unique contexts in the datasets.
|
||||
|
||||
<details>
|
||||
<summary> Code </summary>
|
||||
|
||||
|
||||
```python
|
||||
def extract_unique_contexts(input_directory, output_directory):
|
||||
|
||||
@@ -361,12 +550,12 @@ For the extracted contexts, we insert them into the LightRAG system.
|
||||
|
||||
<details>
|
||||
<summary> Code </summary>
|
||||
|
||||
|
||||
```python
|
||||
def insert_text(rag, file_path):
|
||||
with open(file_path, mode='r') as f:
|
||||
unique_contexts = json.load(f)
|
||||
|
||||
|
||||
retries = 0
|
||||
max_retries = 3
|
||||
while retries < max_retries:
|
||||
@@ -384,11 +573,11 @@ def insert_text(rag, file_path):
|
||||
|
||||
### Step-2 Generate Queries
|
||||
|
||||
We extract tokens from both the first half and the second half of each context in the dataset, then combine them as the dataset description to generate queries.
|
||||
We extract tokens from the first and the second half of each context in the dataset, then combine them as dataset descriptions to generate queries.
|
||||
|
||||
<details>
|
||||
<summary> Code </summary>
|
||||
|
||||
|
||||
```python
|
||||
tokenizer = GPT2Tokenizer.from_pretrained('gpt2')
|
||||
|
||||
@@ -401,7 +590,7 @@ def get_summary(context, tot_tokens=2000):
|
||||
|
||||
summary_tokens = start_tokens + end_tokens
|
||||
summary = tokenizer.convert_tokens_to_string(summary_tokens)
|
||||
|
||||
|
||||
return summary
|
||||
```
|
||||
</details>
|
||||
@@ -411,12 +600,12 @@ For the queries generated in Step-2, we will extract them and query LightRAG.
|
||||
|
||||
<details>
|
||||
<summary> Code </summary>
|
||||
|
||||
|
||||
```python
|
||||
def extract_queries(file_path):
|
||||
with open(file_path, 'r') as f:
|
||||
data = f.read()
|
||||
|
||||
|
||||
data = data.replace('**', '')
|
||||
|
||||
queries = re.findall(r'- Question \d+: (.+)', data)
|
||||
@@ -431,11 +620,16 @@ def extract_queries(file_path):
|
||||
.
|
||||
├── examples
|
||||
│ ├── batch_eval.py
|
||||
│ ├── graph_visual_with_html.py
|
||||
│ ├── graph_visual_with_neo4j.py
|
||||
│ ├── generate_query.py
|
||||
│ ├── lightrag_azure_openai_demo.py
|
||||
│ ├── lightrag_bedrock_demo.py
|
||||
│ ├── lightrag_hf_demo.py
|
||||
│ ├── lightrag_ollama_demo.py
|
||||
│ ├── lightrag_openai_compatible_demo.py
|
||||
│ └── lightrag_openai_demo.py
|
||||
│ ├── lightrag_openai_demo.py
|
||||
│ └── vram_management_demo.py
|
||||
├── lightrag
|
||||
│ ├── __init__.py
|
||||
│ ├── base.py
|
||||
@@ -450,6 +644,8 @@ def extract_queries(file_path):
|
||||
│ ├── Step_1.py
|
||||
│ ├── Step_2.py
|
||||
│ └── Step_3.py
|
||||
├── .gitignore
|
||||
├── .pre-commit-config.yaml
|
||||
├── LICENSE
|
||||
├── README.md
|
||||
├── requirements.txt
|
||||
@@ -470,7 +666,7 @@ def extract_queries(file_path):
|
||||
|
||||
```python
|
||||
@article{guo2024lightrag,
|
||||
title={LightRAG: Simple and Fast Retrieval-Augmented Generation},
|
||||
title={LightRAG: Simple and Fast Retrieval-Augmented Generation},
|
||||
author={Zirui Guo and Lianghao Xia and Yanhua Yu and Tu Ao and Chao Huang},
|
||||
year={2024},
|
||||
eprint={2410.05779},
|
||||
|
||||
@@ -1,4 +1,3 @@
|
||||
import os
|
||||
import re
|
||||
import json
|
||||
import jsonlines
|
||||
@@ -9,28 +8,28 @@ from openai import OpenAI
|
||||
def batch_eval(query_file, result1_file, result2_file, output_file_path):
|
||||
client = OpenAI()
|
||||
|
||||
with open(query_file, 'r') as f:
|
||||
with open(query_file, "r") as f:
|
||||
data = f.read()
|
||||
|
||||
queries = re.findall(r'- Question \d+: (.+)', data)
|
||||
queries = re.findall(r"- Question \d+: (.+)", data)
|
||||
|
||||
with open(result1_file, 'r') as f:
|
||||
with open(result1_file, "r") as f:
|
||||
answers1 = json.load(f)
|
||||
answers1 = [i['result'] for i in answers1]
|
||||
answers1 = [i["result"] for i in answers1]
|
||||
|
||||
with open(result2_file, 'r') as f:
|
||||
with open(result2_file, "r") as f:
|
||||
answers2 = json.load(f)
|
||||
answers2 = [i['result'] for i in answers2]
|
||||
answers2 = [i["result"] for i in answers2]
|
||||
|
||||
requests = []
|
||||
for i, (query, answer1, answer2) in enumerate(zip(queries, answers1, answers2)):
|
||||
sys_prompt = f"""
|
||||
sys_prompt = """
|
||||
---Role---
|
||||
You are an expert tasked with evaluating two answers to the same question based on three criteria: **Comprehensiveness**, **Diversity**, and **Empowerment**.
|
||||
"""
|
||||
|
||||
prompt = f"""
|
||||
You will evaluate two answers to the same question based on three criteria: **Comprehensiveness**, **Diversity**, and **Empowerment**.
|
||||
You will evaluate two answers to the same question based on three criteria: **Comprehensiveness**, **Diversity**, and **Empowerment**.
|
||||
|
||||
- **Comprehensiveness**: How much detail does the answer provide to cover all aspects and details of the question?
|
||||
- **Diversity**: How varied and rich is the answer in providing different perspectives and insights on the question?
|
||||
@@ -69,7 +68,6 @@ def batch_eval(query_file, result1_file, result2_file, output_file_path):
|
||||
}}
|
||||
"""
|
||||
|
||||
|
||||
request_data = {
|
||||
"custom_id": f"request-{i+1}",
|
||||
"method": "POST",
|
||||
@@ -78,22 +76,21 @@ def batch_eval(query_file, result1_file, result2_file, output_file_path):
|
||||
"model": "gpt-4o-mini",
|
||||
"messages": [
|
||||
{"role": "system", "content": sys_prompt},
|
||||
{"role": "user", "content": prompt}
|
||||
{"role": "user", "content": prompt},
|
||||
],
|
||||
}
|
||||
},
|
||||
}
|
||||
|
||||
|
||||
requests.append(request_data)
|
||||
|
||||
with jsonlines.open(output_file_path, mode='w') as writer:
|
||||
with jsonlines.open(output_file_path, mode="w") as writer:
|
||||
for request in requests:
|
||||
writer.write(request)
|
||||
|
||||
print(f"Batch API requests written to {output_file_path}")
|
||||
|
||||
batch_input_file = client.files.create(
|
||||
file=open(output_file_path, "rb"),
|
||||
purpose="batch"
|
||||
file=open(output_file_path, "rb"), purpose="batch"
|
||||
)
|
||||
batch_input_file_id = batch_input_file.id
|
||||
|
||||
@@ -101,12 +98,11 @@ def batch_eval(query_file, result1_file, result2_file, output_file_path):
|
||||
input_file_id=batch_input_file_id,
|
||||
endpoint="/v1/chat/completions",
|
||||
completion_window="24h",
|
||||
metadata={
|
||||
"description": "nightly eval job"
|
||||
}
|
||||
metadata={"description": "nightly eval job"},
|
||||
)
|
||||
|
||||
print(f'Batch {batch.id} has been created.')
|
||||
print(f"Batch {batch.id} has been created.")
|
||||
|
||||
|
||||
if __name__ == "__main__":
|
||||
batch_eval()
|
||||
batch_eval()
|
||||
|
||||
@@ -1,9 +1,8 @@
|
||||
import os
|
||||
|
||||
from openai import OpenAI
|
||||
|
||||
# os.environ["OPENAI_API_KEY"] = ""
|
||||
|
||||
|
||||
def openai_complete_if_cache(
|
||||
model="gpt-4o-mini", prompt=None, system_prompt=None, history_messages=[], **kwargs
|
||||
) -> str:
|
||||
@@ -47,10 +46,10 @@ if __name__ == "__main__":
|
||||
...
|
||||
"""
|
||||
|
||||
result = openai_complete_if_cache(model='gpt-4o-mini', prompt=prompt)
|
||||
result = openai_complete_if_cache(model="gpt-4o-mini", prompt=prompt)
|
||||
|
||||
file_path = f"./queries.txt"
|
||||
file_path = "./queries.txt"
|
||||
with open(file_path, "w") as file:
|
||||
file.write(result)
|
||||
|
||||
print(f"Queries written to {file_path}")
|
||||
print(f"Queries written to {file_path}")
|
||||
|
||||
19
examples/graph_visual_with_html.py
Normal file
19
examples/graph_visual_with_html.py
Normal file
@@ -0,0 +1,19 @@
|
||||
import networkx as nx
|
||||
from pyvis.network import Network
|
||||
import random
|
||||
|
||||
# Load the GraphML file
|
||||
G = nx.read_graphml('./dickens/graph_chunk_entity_relation.graphml')
|
||||
|
||||
# Create a Pyvis network
|
||||
net = Network(notebook=True)
|
||||
|
||||
# Convert NetworkX graph to Pyvis network
|
||||
net.from_nx(G)
|
||||
|
||||
# Add colors to nodes
|
||||
for node in net.nodes:
|
||||
node['color'] = "#{:06x}".format(random.randint(0, 0xFFFFFF))
|
||||
|
||||
# Save and display the network
|
||||
net.show('knowledge_graph.html')
|
||||
118
examples/graph_visual_with_neo4j.py
Normal file
118
examples/graph_visual_with_neo4j.py
Normal file
@@ -0,0 +1,118 @@
|
||||
import os
|
||||
import json
|
||||
from lightrag.utils import xml_to_json
|
||||
from neo4j import GraphDatabase
|
||||
|
||||
# Constants
|
||||
WORKING_DIR = "./dickens"
|
||||
BATCH_SIZE_NODES = 500
|
||||
BATCH_SIZE_EDGES = 100
|
||||
|
||||
# Neo4j connection credentials
|
||||
NEO4J_URI = "bolt://localhost:7687"
|
||||
NEO4J_USERNAME = "neo4j"
|
||||
NEO4J_PASSWORD = "your_password"
|
||||
|
||||
def convert_xml_to_json(xml_path, output_path):
|
||||
"""Converts XML file to JSON and saves the output."""
|
||||
if not os.path.exists(xml_path):
|
||||
print(f"Error: File not found - {xml_path}")
|
||||
return None
|
||||
|
||||
json_data = xml_to_json(xml_path)
|
||||
if json_data:
|
||||
with open(output_path, 'w', encoding='utf-8') as f:
|
||||
json.dump(json_data, f, ensure_ascii=False, indent=2)
|
||||
print(f"JSON file created: {output_path}")
|
||||
return json_data
|
||||
else:
|
||||
print("Failed to create JSON data")
|
||||
return None
|
||||
|
||||
def process_in_batches(tx, query, data, batch_size):
|
||||
"""Process data in batches and execute the given query."""
|
||||
for i in range(0, len(data), batch_size):
|
||||
batch = data[i:i + batch_size]
|
||||
tx.run(query, {"nodes": batch} if "nodes" in query else {"edges": batch})
|
||||
|
||||
def main():
|
||||
# Paths
|
||||
xml_file = os.path.join(WORKING_DIR, 'graph_chunk_entity_relation.graphml')
|
||||
json_file = os.path.join(WORKING_DIR, 'graph_data.json')
|
||||
|
||||
# Convert XML to JSON
|
||||
json_data = convert_xml_to_json(xml_file, json_file)
|
||||
if json_data is None:
|
||||
return
|
||||
|
||||
# Load nodes and edges
|
||||
nodes = json_data.get('nodes', [])
|
||||
edges = json_data.get('edges', [])
|
||||
|
||||
# Neo4j queries
|
||||
create_nodes_query = """
|
||||
UNWIND $nodes AS node
|
||||
MERGE (e:Entity {id: node.id})
|
||||
SET e.entity_type = node.entity_type,
|
||||
e.description = node.description,
|
||||
e.source_id = node.source_id,
|
||||
e.displayName = node.id
|
||||
REMOVE e:Entity
|
||||
WITH e, node
|
||||
CALL apoc.create.addLabels(e, [node.entity_type]) YIELD node AS labeledNode
|
||||
RETURN count(*)
|
||||
"""
|
||||
|
||||
create_edges_query = """
|
||||
UNWIND $edges AS edge
|
||||
MATCH (source {id: edge.source})
|
||||
MATCH (target {id: edge.target})
|
||||
WITH source, target, edge,
|
||||
CASE
|
||||
WHEN edge.keywords CONTAINS 'lead' THEN 'lead'
|
||||
WHEN edge.keywords CONTAINS 'participate' THEN 'participate'
|
||||
WHEN edge.keywords CONTAINS 'uses' THEN 'uses'
|
||||
WHEN edge.keywords CONTAINS 'located' THEN 'located'
|
||||
WHEN edge.keywords CONTAINS 'occurs' THEN 'occurs'
|
||||
ELSE REPLACE(SPLIT(edge.keywords, ',')[0], '\"', '')
|
||||
END AS relType
|
||||
CALL apoc.create.relationship(source, relType, {
|
||||
weight: edge.weight,
|
||||
description: edge.description,
|
||||
keywords: edge.keywords,
|
||||
source_id: edge.source_id
|
||||
}, target) YIELD rel
|
||||
RETURN count(*)
|
||||
"""
|
||||
|
||||
set_displayname_and_labels_query = """
|
||||
MATCH (n)
|
||||
SET n.displayName = n.id
|
||||
WITH n
|
||||
CALL apoc.create.setLabels(n, [n.entity_type]) YIELD node
|
||||
RETURN count(*)
|
||||
"""
|
||||
|
||||
# Create a Neo4j driver
|
||||
driver = GraphDatabase.driver(NEO4J_URI, auth=(NEO4J_USERNAME, NEO4J_PASSWORD))
|
||||
|
||||
try:
|
||||
# Execute queries in batches
|
||||
with driver.session() as session:
|
||||
# Insert nodes in batches
|
||||
session.execute_write(process_in_batches, create_nodes_query, nodes, BATCH_SIZE_NODES)
|
||||
|
||||
# Insert edges in batches
|
||||
session.execute_write(process_in_batches, create_edges_query, edges, BATCH_SIZE_EDGES)
|
||||
|
||||
# Set displayName and labels
|
||||
session.run(set_displayname_and_labels_query)
|
||||
|
||||
except Exception as e:
|
||||
print(f"Error occurred: {e}")
|
||||
|
||||
finally:
|
||||
driver.close()
|
||||
|
||||
if __name__ == "__main__":
|
||||
main()
|
||||
@@ -122,4 +122,4 @@ print("\nResult (Global):")
|
||||
print(rag.query(query_text, param=QueryParam(mode="global")))
|
||||
|
||||
print("\nResult (Hybrid):")
|
||||
print(rag.query(query_text, param=QueryParam(mode="hybrid")))
|
||||
print(rag.query(query_text, param=QueryParam(mode="hybrid")))
|
||||
|
||||
36
examples/lightrag_bedrock_demo.py
Normal file
36
examples/lightrag_bedrock_demo.py
Normal file
@@ -0,0 +1,36 @@
|
||||
"""
|
||||
LightRAG meets Amazon Bedrock ⛰️
|
||||
"""
|
||||
|
||||
import os
|
||||
import logging
|
||||
|
||||
from lightrag import LightRAG, QueryParam
|
||||
from lightrag.llm import bedrock_complete, bedrock_embedding
|
||||
from lightrag.utils import EmbeddingFunc
|
||||
|
||||
logging.getLogger("aiobotocore").setLevel(logging.WARNING)
|
||||
|
||||
WORKING_DIR = "./dickens"
|
||||
if not os.path.exists(WORKING_DIR):
|
||||
os.mkdir(WORKING_DIR)
|
||||
|
||||
rag = LightRAG(
|
||||
working_dir=WORKING_DIR,
|
||||
llm_model_func=bedrock_complete,
|
||||
llm_model_name="Anthropic Claude 3 Haiku // Amazon Bedrock",
|
||||
embedding_func=EmbeddingFunc(
|
||||
embedding_dim=1024, max_token_size=8192, func=bedrock_embedding
|
||||
),
|
||||
)
|
||||
|
||||
with open("./book.txt", "r", encoding="utf-8") as f:
|
||||
rag.insert(f.read())
|
||||
|
||||
for mode in ["naive", "local", "global", "hybrid"]:
|
||||
print("\n+-" + "-" * len(mode) + "-+")
|
||||
print(f"| {mode.capitalize()} |")
|
||||
print("+-" + "-" * len(mode) + "-+\n")
|
||||
print(
|
||||
rag.query("What are the top themes in this story?", param=QueryParam(mode=mode))
|
||||
)
|
||||
@@ -1,10 +1,9 @@
|
||||
import os
|
||||
import sys
|
||||
|
||||
from lightrag import LightRAG, QueryParam
|
||||
from lightrag.llm import hf_model_complete, hf_embedding
|
||||
from lightrag.utils import EmbeddingFunc
|
||||
from transformers import AutoModel,AutoTokenizer
|
||||
from transformers import AutoModel, AutoTokenizer
|
||||
|
||||
WORKING_DIR = "./dickens"
|
||||
|
||||
@@ -13,16 +12,20 @@ if not os.path.exists(WORKING_DIR):
|
||||
|
||||
rag = LightRAG(
|
||||
working_dir=WORKING_DIR,
|
||||
llm_model_func=hf_model_complete,
|
||||
llm_model_name='meta-llama/Llama-3.1-8B-Instruct',
|
||||
llm_model_func=hf_model_complete,
|
||||
llm_model_name="meta-llama/Llama-3.1-8B-Instruct",
|
||||
embedding_func=EmbeddingFunc(
|
||||
embedding_dim=384,
|
||||
max_token_size=5000,
|
||||
func=lambda texts: hf_embedding(
|
||||
texts,
|
||||
tokenizer=AutoTokenizer.from_pretrained("sentence-transformers/all-MiniLM-L6-v2"),
|
||||
embed_model=AutoModel.from_pretrained("sentence-transformers/all-MiniLM-L6-v2")
|
||||
)
|
||||
texts,
|
||||
tokenizer=AutoTokenizer.from_pretrained(
|
||||
"sentence-transformers/all-MiniLM-L6-v2"
|
||||
),
|
||||
embed_model=AutoModel.from_pretrained(
|
||||
"sentence-transformers/all-MiniLM-L6-v2"
|
||||
),
|
||||
),
|
||||
),
|
||||
)
|
||||
|
||||
@@ -31,13 +34,21 @@ with open("./book.txt") as f:
|
||||
rag.insert(f.read())
|
||||
|
||||
# Perform naive search
|
||||
print(rag.query("What are the top themes in this story?", param=QueryParam(mode="naive")))
|
||||
print(
|
||||
rag.query("What are the top themes in this story?", param=QueryParam(mode="naive"))
|
||||
)
|
||||
|
||||
# Perform local search
|
||||
print(rag.query("What are the top themes in this story?", param=QueryParam(mode="local")))
|
||||
print(
|
||||
rag.query("What are the top themes in this story?", param=QueryParam(mode="local"))
|
||||
)
|
||||
|
||||
# Perform global search
|
||||
print(rag.query("What are the top themes in this story?", param=QueryParam(mode="global")))
|
||||
print(
|
||||
rag.query("What are the top themes in this story?", param=QueryParam(mode="global"))
|
||||
)
|
||||
|
||||
# Perform hybrid search
|
||||
print(rag.query("What are the top themes in this story?", param=QueryParam(mode="hybrid")))
|
||||
print(
|
||||
rag.query("What are the top themes in this story?", param=QueryParam(mode="hybrid"))
|
||||
)
|
||||
|
||||
@@ -6,10 +6,11 @@ from lightrag.utils import EmbeddingFunc
|
||||
import numpy as np
|
||||
|
||||
WORKING_DIR = "./dickens"
|
||||
|
||||
|
||||
if not os.path.exists(WORKING_DIR):
|
||||
os.mkdir(WORKING_DIR)
|
||||
|
||||
|
||||
async def llm_model_func(
|
||||
prompt, system_prompt=None, history_messages=[], **kwargs
|
||||
) -> str:
|
||||
@@ -20,17 +21,19 @@ async def llm_model_func(
|
||||
history_messages=history_messages,
|
||||
api_key=os.getenv("UPSTAGE_API_KEY"),
|
||||
base_url="https://api.upstage.ai/v1/solar",
|
||||
**kwargs
|
||||
**kwargs,
|
||||
)
|
||||
|
||||
|
||||
async def embedding_func(texts: list[str]) -> np.ndarray:
|
||||
return await openai_embedding(
|
||||
texts,
|
||||
model="solar-embedding-1-large-query",
|
||||
api_key=os.getenv("UPSTAGE_API_KEY"),
|
||||
base_url="https://api.upstage.ai/v1/solar"
|
||||
base_url="https://api.upstage.ai/v1/solar",
|
||||
)
|
||||
|
||||
|
||||
# function test
|
||||
async def test_funcs():
|
||||
result = await llm_model_func("How are you?")
|
||||
@@ -39,6 +42,7 @@ async def test_funcs():
|
||||
result = await embedding_func(["How are you?"])
|
||||
print("embedding_func: ", result)
|
||||
|
||||
|
||||
asyncio.run(test_funcs())
|
||||
|
||||
|
||||
@@ -46,10 +50,8 @@ rag = LightRAG(
|
||||
working_dir=WORKING_DIR,
|
||||
llm_model_func=llm_model_func,
|
||||
embedding_func=EmbeddingFunc(
|
||||
embedding_dim=4096,
|
||||
max_token_size=8192,
|
||||
func=embedding_func
|
||||
)
|
||||
embedding_dim=4096, max_token_size=8192, func=embedding_func
|
||||
),
|
||||
)
|
||||
|
||||
|
||||
@@ -57,13 +59,21 @@ with open("./book.txt") as f:
|
||||
rag.insert(f.read())
|
||||
|
||||
# Perform naive search
|
||||
print(rag.query("What are the top themes in this story?", param=QueryParam(mode="naive")))
|
||||
print(
|
||||
rag.query("What are the top themes in this story?", param=QueryParam(mode="naive"))
|
||||
)
|
||||
|
||||
# Perform local search
|
||||
print(rag.query("What are the top themes in this story?", param=QueryParam(mode="local")))
|
||||
print(
|
||||
rag.query("What are the top themes in this story?", param=QueryParam(mode="local"))
|
||||
)
|
||||
|
||||
# Perform global search
|
||||
print(rag.query("What are the top themes in this story?", param=QueryParam(mode="global")))
|
||||
print(
|
||||
rag.query("What are the top themes in this story?", param=QueryParam(mode="global"))
|
||||
)
|
||||
|
||||
# Perform hybrid search
|
||||
print(rag.query("What are the top themes in this story?", param=QueryParam(mode="hybrid")))
|
||||
print(
|
||||
rag.query("What are the top themes in this story?", param=QueryParam(mode="hybrid"))
|
||||
)
|
||||
|
||||
@@ -1,9 +1,7 @@
|
||||
import os
|
||||
import sys
|
||||
|
||||
from lightrag import LightRAG, QueryParam
|
||||
from lightrag.llm import gpt_4o_mini_complete, gpt_4o_complete
|
||||
from transformers import AutoModel,AutoTokenizer
|
||||
from lightrag.llm import gpt_4o_mini_complete
|
||||
|
||||
WORKING_DIR = "./dickens"
|
||||
|
||||
@@ -12,7 +10,7 @@ if not os.path.exists(WORKING_DIR):
|
||||
|
||||
rag = LightRAG(
|
||||
working_dir=WORKING_DIR,
|
||||
llm_model_func=gpt_4o_mini_complete
|
||||
llm_model_func=gpt_4o_mini_complete,
|
||||
# llm_model_func=gpt_4o_complete
|
||||
)
|
||||
|
||||
@@ -21,13 +19,21 @@ with open("./book.txt") as f:
|
||||
rag.insert(f.read())
|
||||
|
||||
# Perform naive search
|
||||
print(rag.query("What are the top themes in this story?", param=QueryParam(mode="naive")))
|
||||
print(
|
||||
rag.query("What are the top themes in this story?", param=QueryParam(mode="naive"))
|
||||
)
|
||||
|
||||
# Perform local search
|
||||
print(rag.query("What are the top themes in this story?", param=QueryParam(mode="local")))
|
||||
print(
|
||||
rag.query("What are the top themes in this story?", param=QueryParam(mode="local"))
|
||||
)
|
||||
|
||||
# Perform global search
|
||||
print(rag.query("What are the top themes in this story?", param=QueryParam(mode="global")))
|
||||
print(
|
||||
rag.query("What are the top themes in this story?", param=QueryParam(mode="global"))
|
||||
)
|
||||
|
||||
# Perform hybrid search
|
||||
print(rag.query("What are the top themes in this story?", param=QueryParam(mode="hybrid")))
|
||||
print(
|
||||
rag.query("What are the top themes in this story?", param=QueryParam(mode="hybrid"))
|
||||
)
|
||||
|
||||
82
examples/vram_management_demo.py
Normal file
82
examples/vram_management_demo.py
Normal file
@@ -0,0 +1,82 @@
|
||||
import os
|
||||
import time
|
||||
from lightrag import LightRAG, QueryParam
|
||||
from lightrag.llm import ollama_model_complete, ollama_embedding
|
||||
from lightrag.utils import EmbeddingFunc
|
||||
|
||||
# Working directory and the directory path for text files
|
||||
WORKING_DIR = "./dickens"
|
||||
TEXT_FILES_DIR = "/llm/mt"
|
||||
|
||||
# Create the working directory if it doesn't exist
|
||||
if not os.path.exists(WORKING_DIR):
|
||||
os.mkdir(WORKING_DIR)
|
||||
|
||||
# Initialize LightRAG
|
||||
rag = LightRAG(
|
||||
working_dir=WORKING_DIR,
|
||||
llm_model_func=ollama_model_complete,
|
||||
llm_model_name="qwen2.5:3b-instruct-max-context",
|
||||
embedding_func=EmbeddingFunc(
|
||||
embedding_dim=768,
|
||||
max_token_size=8192,
|
||||
func=lambda texts: ollama_embedding(texts, embed_model="nomic-embed-text"),
|
||||
),
|
||||
)
|
||||
|
||||
# Read all .txt files from the TEXT_FILES_DIR directory
|
||||
texts = []
|
||||
for filename in os.listdir(TEXT_FILES_DIR):
|
||||
if filename.endswith('.txt'):
|
||||
file_path = os.path.join(TEXT_FILES_DIR, filename)
|
||||
with open(file_path, 'r', encoding='utf-8') as file:
|
||||
texts.append(file.read())
|
||||
|
||||
# Batch insert texts into LightRAG with a retry mechanism
|
||||
def insert_texts_with_retry(rag, texts, retries=3, delay=5):
|
||||
for _ in range(retries):
|
||||
try:
|
||||
rag.insert(texts)
|
||||
return
|
||||
except Exception as e:
|
||||
print(f"Error occurred during insertion: {e}. Retrying in {delay} seconds...")
|
||||
time.sleep(delay)
|
||||
raise RuntimeError("Failed to insert texts after multiple retries.")
|
||||
|
||||
insert_texts_with_retry(rag, texts)
|
||||
|
||||
# Perform different types of queries and handle potential errors
|
||||
try:
|
||||
print(rag.query("What are the top themes in this story?", param=QueryParam(mode="naive")))
|
||||
except Exception as e:
|
||||
print(f"Error performing naive search: {e}")
|
||||
|
||||
try:
|
||||
print(rag.query("What are the top themes in this story?", param=QueryParam(mode="local")))
|
||||
except Exception as e:
|
||||
print(f"Error performing local search: {e}")
|
||||
|
||||
try:
|
||||
print(rag.query("What are the top themes in this story?", param=QueryParam(mode="global")))
|
||||
except Exception as e:
|
||||
print(f"Error performing global search: {e}")
|
||||
|
||||
try:
|
||||
print(rag.query("What are the top themes in this story?", param=QueryParam(mode="hybrid")))
|
||||
except Exception as e:
|
||||
print(f"Error performing hybrid search: {e}")
|
||||
|
||||
# Function to clear VRAM resources
|
||||
def clear_vram():
|
||||
os.system("sudo nvidia-smi --gpu-reset")
|
||||
|
||||
# Regularly clear VRAM to prevent overflow
|
||||
clear_vram_interval = 3600 # Clear once every hour
|
||||
start_time = time.time()
|
||||
|
||||
while True:
|
||||
current_time = time.time()
|
||||
if current_time - start_time > clear_vram_interval:
|
||||
clear_vram()
|
||||
start_time = current_time
|
||||
time.sleep(60) # Check the time every minute
|
||||
@@ -1,5 +1,5 @@
|
||||
from .lightrag import LightRAG, QueryParam
|
||||
from .lightrag import LightRAG as LightRAG, QueryParam as QueryParam
|
||||
|
||||
__version__ = "0.0.6"
|
||||
__version__ = "0.0.7"
|
||||
__author__ = "Zirui Guo"
|
||||
__url__ = "https://github.com/HKUDS/LightRAG"
|
||||
|
||||
@@ -12,15 +12,16 @@ TextChunkSchema = TypedDict(
|
||||
|
||||
T = TypeVar("T")
|
||||
|
||||
|
||||
@dataclass
|
||||
class QueryParam:
|
||||
mode: Literal["local", "global", "hybrid", "naive"] = "global"
|
||||
only_need_context: bool = False
|
||||
response_type: str = "Multiple Paragraphs"
|
||||
top_k: int = 60
|
||||
max_token_for_text_unit: int = 4000
|
||||
max_token_for_text_unit: int = 4000
|
||||
max_token_for_global_context: int = 4000
|
||||
max_token_for_local_context: int = 4000
|
||||
max_token_for_local_context: int = 4000
|
||||
|
||||
|
||||
@dataclass
|
||||
@@ -36,6 +37,7 @@ class StorageNameSpace:
|
||||
"""commit the storage operations after querying"""
|
||||
pass
|
||||
|
||||
|
||||
@dataclass
|
||||
class BaseVectorStorage(StorageNameSpace):
|
||||
embedding_func: EmbeddingFunc
|
||||
@@ -50,6 +52,7 @@ class BaseVectorStorage(StorageNameSpace):
|
||||
"""
|
||||
raise NotImplementedError
|
||||
|
||||
|
||||
@dataclass
|
||||
class BaseKVStorage(Generic[T], StorageNameSpace):
|
||||
async def all_keys(self) -> list[str]:
|
||||
@@ -72,7 +75,7 @@ class BaseKVStorage(Generic[T], StorageNameSpace):
|
||||
|
||||
async def drop(self):
|
||||
raise NotImplementedError
|
||||
|
||||
|
||||
|
||||
@dataclass
|
||||
class BaseGraphStorage(StorageNameSpace):
|
||||
@@ -113,4 +116,4 @@ class BaseGraphStorage(StorageNameSpace):
|
||||
raise NotImplementedError
|
||||
|
||||
async def embed_nodes(self, algorithm: str) -> tuple[np.ndarray, list[str]]:
|
||||
raise NotImplementedError("Node embedding is not used in lightrag.")
|
||||
raise NotImplementedError("Node embedding is not used in lightrag.")
|
||||
|
||||
@@ -3,10 +3,12 @@ import os
|
||||
from dataclasses import asdict, dataclass, field
|
||||
from datetime import datetime
|
||||
from functools import partial
|
||||
from typing import Type, cast, Any
|
||||
from transformers import AutoModel,AutoTokenizer, AutoModelForCausalLM
|
||||
from typing import Type, cast
|
||||
|
||||
from .llm import gpt_4o_complete, gpt_4o_mini_complete, openai_embedding, hf_model_complete, hf_embedding
|
||||
from .llm import (
|
||||
gpt_4o_mini_complete,
|
||||
openai_embedding,
|
||||
)
|
||||
from .operate import (
|
||||
chunking_by_token_size,
|
||||
extract_entities,
|
||||
@@ -37,6 +39,7 @@ from .base import (
|
||||
QueryParam,
|
||||
)
|
||||
|
||||
|
||||
def always_get_an_event_loop() -> asyncio.AbstractEventLoop:
|
||||
try:
|
||||
loop = asyncio.get_running_loop()
|
||||
@@ -69,7 +72,6 @@ class LightRAG:
|
||||
"dimensions": 1536,
|
||||
"num_walks": 10,
|
||||
"walk_length": 40,
|
||||
"num_walks": 10,
|
||||
"window_size": 2,
|
||||
"iterations": 3,
|
||||
"random_seed": 3,
|
||||
@@ -77,13 +79,13 @@ class LightRAG:
|
||||
)
|
||||
|
||||
# embedding_func: EmbeddingFunc = field(default_factory=lambda:hf_embedding)
|
||||
embedding_func: EmbeddingFunc = field(default_factory=lambda:openai_embedding)
|
||||
embedding_func: EmbeddingFunc = field(default_factory=lambda: openai_embedding)
|
||||
embedding_batch_num: int = 32
|
||||
embedding_func_max_async: int = 16
|
||||
|
||||
# LLM
|
||||
llm_model_func: callable = gpt_4o_mini_complete#hf_model_complete#
|
||||
llm_model_name: str = 'meta-llama/Llama-3.2-1B-Instruct'#'meta-llama/Llama-3.2-1B'#'google/gemma-2-2b-it'
|
||||
llm_model_func: callable = gpt_4o_mini_complete # hf_model_complete#
|
||||
llm_model_name: str = "meta-llama/Llama-3.2-1B-Instruct" #'meta-llama/Llama-3.2-1B'#'google/gemma-2-2b-it'
|
||||
llm_model_max_token_size: int = 32768
|
||||
llm_model_max_async: int = 16
|
||||
llm_model_kwargs: dict = field(default_factory=dict)
|
||||
@@ -99,11 +101,11 @@ class LightRAG:
|
||||
addon_params: dict = field(default_factory=dict)
|
||||
convert_response_to_json_func: callable = convert_response_to_json
|
||||
|
||||
def __post_init__(self):
|
||||
def __post_init__(self):
|
||||
log_file = os.path.join(self.working_dir, "lightrag.log")
|
||||
set_logger(log_file)
|
||||
logger.info(f"Logger initialized for working directory: {self.working_dir}")
|
||||
|
||||
|
||||
_print_config = ",\n ".join([f"{k} = {v}" for k, v in asdict(self).items()])
|
||||
logger.debug(f"LightRAG init with param:\n {_print_config}\n")
|
||||
|
||||
@@ -134,30 +136,24 @@ class LightRAG:
|
||||
self.embedding_func
|
||||
)
|
||||
|
||||
self.entities_vdb = (
|
||||
self.vector_db_storage_cls(
|
||||
namespace="entities",
|
||||
global_config=asdict(self),
|
||||
embedding_func=self.embedding_func,
|
||||
meta_fields={"entity_name"}
|
||||
)
|
||||
self.entities_vdb = self.vector_db_storage_cls(
|
||||
namespace="entities",
|
||||
global_config=asdict(self),
|
||||
embedding_func=self.embedding_func,
|
||||
meta_fields={"entity_name"},
|
||||
)
|
||||
self.relationships_vdb = (
|
||||
self.vector_db_storage_cls(
|
||||
namespace="relationships",
|
||||
global_config=asdict(self),
|
||||
embedding_func=self.embedding_func,
|
||||
meta_fields={"src_id", "tgt_id"}
|
||||
)
|
||||
self.relationships_vdb = self.vector_db_storage_cls(
|
||||
namespace="relationships",
|
||||
global_config=asdict(self),
|
||||
embedding_func=self.embedding_func,
|
||||
meta_fields={"src_id", "tgt_id"},
|
||||
)
|
||||
self.chunks_vdb = (
|
||||
self.vector_db_storage_cls(
|
||||
namespace="chunks",
|
||||
global_config=asdict(self),
|
||||
embedding_func=self.embedding_func,
|
||||
)
|
||||
self.chunks_vdb = self.vector_db_storage_cls(
|
||||
namespace="chunks",
|
||||
global_config=asdict(self),
|
||||
embedding_func=self.embedding_func,
|
||||
)
|
||||
|
||||
|
||||
self.llm_model_func = limit_async_func_call(self.llm_model_max_async)(
|
||||
partial(self.llm_model_func, hashing_kv=self.llm_response_cache, **self.llm_model_kwargs)
|
||||
)
|
||||
@@ -178,7 +174,7 @@ class LightRAG:
|
||||
_add_doc_keys = await self.full_docs.filter_keys(list(new_docs.keys()))
|
||||
new_docs = {k: v for k, v in new_docs.items() if k in _add_doc_keys}
|
||||
if not len(new_docs):
|
||||
logger.warning(f"All docs are already in the storage")
|
||||
logger.warning("All docs are already in the storage")
|
||||
return
|
||||
logger.info(f"[New Docs] inserting {len(new_docs)} docs")
|
||||
|
||||
@@ -204,7 +200,7 @@ class LightRAG:
|
||||
k: v for k, v in inserting_chunks.items() if k in _add_chunk_keys
|
||||
}
|
||||
if not len(inserting_chunks):
|
||||
logger.warning(f"All chunks are already in the storage")
|
||||
logger.warning("All chunks are already in the storage")
|
||||
return
|
||||
logger.info(f"[New Chunks] inserting {len(inserting_chunks)} chunks")
|
||||
|
||||
@@ -247,7 +243,7 @@ class LightRAG:
|
||||
def query(self, query: str, param: QueryParam = QueryParam()):
|
||||
loop = always_get_an_event_loop()
|
||||
return loop.run_until_complete(self.aquery(query, param))
|
||||
|
||||
|
||||
async def aquery(self, query: str, param: QueryParam = QueryParam()):
|
||||
if param.mode == "local":
|
||||
response = await local_query(
|
||||
@@ -291,7 +287,6 @@ class LightRAG:
|
||||
raise ValueError(f"Unknown mode {param.mode}")
|
||||
await self._query_done()
|
||||
return response
|
||||
|
||||
|
||||
async def _query_done(self):
|
||||
tasks = []
|
||||
@@ -300,5 +295,3 @@ class LightRAG:
|
||||
continue
|
||||
tasks.append(cast(StorageNameSpace, storage_inst).index_done_callback())
|
||||
await asyncio.gather(*tasks)
|
||||
|
||||
|
||||
|
||||
277
lightrag/llm.py
277
lightrag/llm.py
@@ -1,4 +1,7 @@
|
||||
import os
|
||||
import copy
|
||||
import json
|
||||
import aioboto3
|
||||
import numpy as np
|
||||
import ollama
|
||||
from openai import AsyncOpenAI, APIConnectionError, RateLimitError, Timeout
|
||||
@@ -8,24 +11,34 @@ from tenacity import (
|
||||
wait_exponential,
|
||||
retry_if_exception_type,
|
||||
)
|
||||
from transformers import AutoModel,AutoTokenizer, AutoModelForCausalLM
|
||||
from transformers import AutoTokenizer, AutoModelForCausalLM
|
||||
import torch
|
||||
from .base import BaseKVStorage
|
||||
from .utils import compute_args_hash, wrap_embedding_func_with_attrs
|
||||
import copy
|
||||
|
||||
os.environ["TOKENIZERS_PARALLELISM"] = "false"
|
||||
|
||||
|
||||
@retry(
|
||||
stop=stop_after_attempt(3),
|
||||
wait=wait_exponential(multiplier=1, min=4, max=10),
|
||||
retry=retry_if_exception_type((RateLimitError, APIConnectionError, Timeout)),
|
||||
)
|
||||
async def openai_complete_if_cache(
|
||||
model, prompt, system_prompt=None, history_messages=[], base_url=None, api_key=None, **kwargs
|
||||
model,
|
||||
prompt,
|
||||
system_prompt=None,
|
||||
history_messages=[],
|
||||
base_url=None,
|
||||
api_key=None,
|
||||
**kwargs,
|
||||
) -> str:
|
||||
if api_key:
|
||||
os.environ["OPENAI_API_KEY"] = api_key
|
||||
|
||||
openai_async_client = AsyncOpenAI() if base_url is None else AsyncOpenAI(base_url=base_url)
|
||||
openai_async_client = (
|
||||
AsyncOpenAI() if base_url is None else AsyncOpenAI(base_url=base_url)
|
||||
)
|
||||
hashing_kv: BaseKVStorage = kwargs.pop("hashing_kv", None)
|
||||
messages = []
|
||||
if system_prompt:
|
||||
@@ -48,15 +61,105 @@ async def openai_complete_if_cache(
|
||||
)
|
||||
return response.choices[0].message.content
|
||||
|
||||
|
||||
class BedrockError(Exception):
|
||||
"""Generic error for issues related to Amazon Bedrock"""
|
||||
|
||||
|
||||
@retry(
|
||||
stop=stop_after_attempt(5),
|
||||
wait=wait_exponential(multiplier=1, max=60),
|
||||
retry=retry_if_exception_type((BedrockError)),
|
||||
)
|
||||
async def bedrock_complete_if_cache(
|
||||
model,
|
||||
prompt,
|
||||
system_prompt=None,
|
||||
history_messages=[],
|
||||
aws_access_key_id=None,
|
||||
aws_secret_access_key=None,
|
||||
aws_session_token=None,
|
||||
**kwargs,
|
||||
) -> str:
|
||||
os.environ["AWS_ACCESS_KEY_ID"] = os.environ.get(
|
||||
"AWS_ACCESS_KEY_ID", aws_access_key_id
|
||||
)
|
||||
os.environ["AWS_SECRET_ACCESS_KEY"] = os.environ.get(
|
||||
"AWS_SECRET_ACCESS_KEY", aws_secret_access_key
|
||||
)
|
||||
os.environ["AWS_SESSION_TOKEN"] = os.environ.get(
|
||||
"AWS_SESSION_TOKEN", aws_session_token
|
||||
)
|
||||
|
||||
# Fix message history format
|
||||
messages = []
|
||||
for history_message in history_messages:
|
||||
message = copy.copy(history_message)
|
||||
message["content"] = [{"text": message["content"]}]
|
||||
messages.append(message)
|
||||
|
||||
# Add user prompt
|
||||
messages.append({"role": "user", "content": [{"text": prompt}]})
|
||||
|
||||
# Initialize Converse API arguments
|
||||
args = {"modelId": model, "messages": messages}
|
||||
|
||||
# Define system prompt
|
||||
if system_prompt:
|
||||
args["system"] = [{"text": system_prompt}]
|
||||
|
||||
# Map and set up inference parameters
|
||||
inference_params_map = {
|
||||
"max_tokens": "maxTokens",
|
||||
"top_p": "topP",
|
||||
"stop_sequences": "stopSequences",
|
||||
}
|
||||
if inference_params := list(
|
||||
set(kwargs) & set(["max_tokens", "temperature", "top_p", "stop_sequences"])
|
||||
):
|
||||
args["inferenceConfig"] = {}
|
||||
for param in inference_params:
|
||||
args["inferenceConfig"][inference_params_map.get(param, param)] = (
|
||||
kwargs.pop(param)
|
||||
)
|
||||
|
||||
hashing_kv: BaseKVStorage = kwargs.pop("hashing_kv", None)
|
||||
if hashing_kv is not None:
|
||||
args_hash = compute_args_hash(model, messages)
|
||||
if_cache_return = await hashing_kv.get_by_id(args_hash)
|
||||
if if_cache_return is not None:
|
||||
return if_cache_return["return"]
|
||||
|
||||
# Call model via Converse API
|
||||
session = aioboto3.Session()
|
||||
async with session.client("bedrock-runtime") as bedrock_async_client:
|
||||
try:
|
||||
response = await bedrock_async_client.converse(**args, **kwargs)
|
||||
except Exception as e:
|
||||
raise BedrockError(e)
|
||||
|
||||
if hashing_kv is not None:
|
||||
await hashing_kv.upsert(
|
||||
{
|
||||
args_hash: {
|
||||
"return": response["output"]["message"]["content"][0]["text"],
|
||||
"model": model,
|
||||
}
|
||||
}
|
||||
)
|
||||
|
||||
return response["output"]["message"]["content"][0]["text"]
|
||||
|
||||
|
||||
async def hf_model_if_cache(
|
||||
model, prompt, system_prompt=None, history_messages=[], **kwargs
|
||||
) -> str:
|
||||
model_name = model
|
||||
hf_tokenizer = AutoTokenizer.from_pretrained(model_name,device_map = 'auto')
|
||||
if hf_tokenizer.pad_token == None:
|
||||
hf_tokenizer = AutoTokenizer.from_pretrained(model_name, device_map="auto")
|
||||
if hf_tokenizer.pad_token is None:
|
||||
# print("use eos token")
|
||||
hf_tokenizer.pad_token = hf_tokenizer.eos_token
|
||||
hf_model = AutoModelForCausalLM.from_pretrained(model_name,device_map = 'auto')
|
||||
hf_model = AutoModelForCausalLM.from_pretrained(model_name, device_map="auto")
|
||||
hashing_kv: BaseKVStorage = kwargs.pop("hashing_kv", None)
|
||||
messages = []
|
||||
if system_prompt:
|
||||
@@ -69,30 +172,51 @@ async def hf_model_if_cache(
|
||||
if_cache_return = await hashing_kv.get_by_id(args_hash)
|
||||
if if_cache_return is not None:
|
||||
return if_cache_return["return"]
|
||||
input_prompt = ''
|
||||
input_prompt = ""
|
||||
try:
|
||||
input_prompt = hf_tokenizer.apply_chat_template(messages, tokenize=False, add_generation_prompt=True)
|
||||
except:
|
||||
input_prompt = hf_tokenizer.apply_chat_template(
|
||||
messages, tokenize=False, add_generation_prompt=True
|
||||
)
|
||||
except Exception:
|
||||
try:
|
||||
ori_message = copy.deepcopy(messages)
|
||||
if messages[0]['role'] == "system":
|
||||
messages[1]['content'] = "<system>" + messages[0]['content'] + "</system>\n" + messages[1]['content']
|
||||
if messages[0]["role"] == "system":
|
||||
messages[1]["content"] = (
|
||||
"<system>"
|
||||
+ messages[0]["content"]
|
||||
+ "</system>\n"
|
||||
+ messages[1]["content"]
|
||||
)
|
||||
messages = messages[1:]
|
||||
input_prompt = hf_tokenizer.apply_chat_template(messages, tokenize=False, add_generation_prompt=True)
|
||||
except:
|
||||
input_prompt = hf_tokenizer.apply_chat_template(
|
||||
messages, tokenize=False, add_generation_prompt=True
|
||||
)
|
||||
except Exception:
|
||||
len_message = len(ori_message)
|
||||
for msgid in range(len_message):
|
||||
input_prompt =input_prompt+ '<'+ori_message[msgid]['role']+'>'+ori_message[msgid]['content']+'</'+ori_message[msgid]['role']+'>\n'
|
||||
|
||||
input_ids = hf_tokenizer(input_prompt, return_tensors='pt', padding=True, truncation=True).to("cuda")
|
||||
output = hf_model.generate(**input_ids, max_new_tokens=200, num_return_sequences=1,early_stopping = True)
|
||||
input_prompt = (
|
||||
input_prompt
|
||||
+ "<"
|
||||
+ ori_message[msgid]["role"]
|
||||
+ ">"
|
||||
+ ori_message[msgid]["content"]
|
||||
+ "</"
|
||||
+ ori_message[msgid]["role"]
|
||||
+ ">\n"
|
||||
)
|
||||
|
||||
input_ids = hf_tokenizer(
|
||||
input_prompt, return_tensors="pt", padding=True, truncation=True
|
||||
).to("cuda")
|
||||
output = hf_model.generate(
|
||||
**input_ids, max_new_tokens=200, num_return_sequences=1, early_stopping=True
|
||||
)
|
||||
response_text = hf_tokenizer.decode(output[0], skip_special_tokens=True)
|
||||
if hashing_kv is not None:
|
||||
await hashing_kv.upsert(
|
||||
{args_hash: {"return": response_text, "model": model}}
|
||||
)
|
||||
await hashing_kv.upsert({args_hash: {"return": response_text, "model": model}})
|
||||
return response_text
|
||||
|
||||
|
||||
async def ollama_model_if_cache(
|
||||
model, prompt, system_prompt=None, history_messages=[], **kwargs
|
||||
) -> str:
|
||||
@@ -124,6 +248,7 @@ async def ollama_model_if_cache(
|
||||
|
||||
return result
|
||||
|
||||
|
||||
async def gpt_4o_complete(
|
||||
prompt, system_prompt=None, history_messages=[], **kwargs
|
||||
) -> str:
|
||||
@@ -147,10 +272,23 @@ async def gpt_4o_mini_complete(
|
||||
**kwargs,
|
||||
)
|
||||
|
||||
|
||||
async def bedrock_complete(
|
||||
prompt, system_prompt=None, history_messages=[], **kwargs
|
||||
) -> str:
|
||||
return await bedrock_complete_if_cache(
|
||||
"anthropic.claude-3-haiku-20240307-v1:0",
|
||||
prompt,
|
||||
system_prompt=system_prompt,
|
||||
history_messages=history_messages,
|
||||
**kwargs,
|
||||
)
|
||||
|
||||
|
||||
async def hf_model_complete(
|
||||
prompt, system_prompt=None, history_messages=[], **kwargs
|
||||
) -> str:
|
||||
model_name = kwargs['hashing_kv'].global_config['llm_model_name']
|
||||
model_name = kwargs["hashing_kv"].global_config["llm_model_name"]
|
||||
return await hf_model_if_cache(
|
||||
model_name,
|
||||
prompt,
|
||||
@@ -159,10 +297,11 @@ async def hf_model_complete(
|
||||
**kwargs,
|
||||
)
|
||||
|
||||
|
||||
async def ollama_model_complete(
|
||||
prompt, system_prompt=None, history_messages=[], **kwargs
|
||||
) -> str:
|
||||
model_name = kwargs['hashing_kv'].global_config['llm_model_name']
|
||||
model_name = kwargs["hashing_kv"].global_config["llm_model_name"]
|
||||
return await ollama_model_if_cache(
|
||||
model_name,
|
||||
prompt,
|
||||
@@ -171,30 +310,113 @@ async def ollama_model_complete(
|
||||
**kwargs,
|
||||
)
|
||||
|
||||
|
||||
@wrap_embedding_func_with_attrs(embedding_dim=1536, max_token_size=8192)
|
||||
@retry(
|
||||
stop=stop_after_attempt(3),
|
||||
wait=wait_exponential(multiplier=1, min=4, max=10),
|
||||
retry=retry_if_exception_type((RateLimitError, APIConnectionError, Timeout)),
|
||||
)
|
||||
async def openai_embedding(texts: list[str], model: str = "text-embedding-3-small", base_url: str = None, api_key: str = None) -> np.ndarray:
|
||||
async def openai_embedding(
|
||||
texts: list[str],
|
||||
model: str = "text-embedding-3-small",
|
||||
base_url: str = None,
|
||||
api_key: str = None,
|
||||
) -> np.ndarray:
|
||||
if api_key:
|
||||
os.environ["OPENAI_API_KEY"] = api_key
|
||||
|
||||
openai_async_client = AsyncOpenAI() if base_url is None else AsyncOpenAI(base_url=base_url)
|
||||
openai_async_client = (
|
||||
AsyncOpenAI() if base_url is None else AsyncOpenAI(base_url=base_url)
|
||||
)
|
||||
response = await openai_async_client.embeddings.create(
|
||||
model=model, input=texts, encoding_format="float"
|
||||
)
|
||||
return np.array([dp.embedding for dp in response.data])
|
||||
|
||||
|
||||
# @wrap_embedding_func_with_attrs(embedding_dim=1024, max_token_size=8192)
|
||||
# @retry(
|
||||
# stop=stop_after_attempt(3),
|
||||
# wait=wait_exponential(multiplier=1, min=4, max=10),
|
||||
# retry=retry_if_exception_type((RateLimitError, APIConnectionError, Timeout)), # TODO: fix exceptions
|
||||
# )
|
||||
async def bedrock_embedding(
|
||||
texts: list[str],
|
||||
model: str = "amazon.titan-embed-text-v2:0",
|
||||
aws_access_key_id=None,
|
||||
aws_secret_access_key=None,
|
||||
aws_session_token=None,
|
||||
) -> np.ndarray:
|
||||
os.environ["AWS_ACCESS_KEY_ID"] = os.environ.get(
|
||||
"AWS_ACCESS_KEY_ID", aws_access_key_id
|
||||
)
|
||||
os.environ["AWS_SECRET_ACCESS_KEY"] = os.environ.get(
|
||||
"AWS_SECRET_ACCESS_KEY", aws_secret_access_key
|
||||
)
|
||||
os.environ["AWS_SESSION_TOKEN"] = os.environ.get(
|
||||
"AWS_SESSION_TOKEN", aws_session_token
|
||||
)
|
||||
|
||||
session = aioboto3.Session()
|
||||
async with session.client("bedrock-runtime") as bedrock_async_client:
|
||||
if (model_provider := model.split(".")[0]) == "amazon":
|
||||
embed_texts = []
|
||||
for text in texts:
|
||||
if "v2" in model:
|
||||
body = json.dumps(
|
||||
{
|
||||
"inputText": text,
|
||||
# 'dimensions': embedding_dim,
|
||||
"embeddingTypes": ["float"],
|
||||
}
|
||||
)
|
||||
elif "v1" in model:
|
||||
body = json.dumps({"inputText": text})
|
||||
else:
|
||||
raise ValueError(f"Model {model} is not supported!")
|
||||
|
||||
response = await bedrock_async_client.invoke_model(
|
||||
modelId=model,
|
||||
body=body,
|
||||
accept="application/json",
|
||||
contentType="application/json",
|
||||
)
|
||||
|
||||
response_body = await response.get("body").json()
|
||||
|
||||
embed_texts.append(response_body["embedding"])
|
||||
elif model_provider == "cohere":
|
||||
body = json.dumps(
|
||||
{"texts": texts, "input_type": "search_document", "truncate": "NONE"}
|
||||
)
|
||||
|
||||
response = await bedrock_async_client.invoke_model(
|
||||
model=model,
|
||||
body=body,
|
||||
accept="application/json",
|
||||
contentType="application/json",
|
||||
)
|
||||
|
||||
response_body = json.loads(response.get("body").read())
|
||||
|
||||
embed_texts = response_body["embeddings"]
|
||||
else:
|
||||
raise ValueError(f"Model provider '{model_provider}' is not supported!")
|
||||
|
||||
return np.array(embed_texts)
|
||||
|
||||
|
||||
async def hf_embedding(texts: list[str], tokenizer, embed_model) -> np.ndarray:
|
||||
input_ids = tokenizer(texts, return_tensors='pt', padding=True, truncation=True).input_ids
|
||||
input_ids = tokenizer(
|
||||
texts, return_tensors="pt", padding=True, truncation=True
|
||||
).input_ids
|
||||
with torch.no_grad():
|
||||
outputs = embed_model(input_ids)
|
||||
embeddings = outputs.last_hidden_state.mean(dim=1)
|
||||
return embeddings.detach().numpy()
|
||||
|
||||
|
||||
async def ollama_embedding(texts: list[str], embed_model, **kwargs) -> np.ndarray:
|
||||
embed_text = []
|
||||
ollama_client = ollama.Client(**kwargs)
|
||||
@@ -204,11 +426,12 @@ async def ollama_embedding(texts: list[str], embed_model, **kwargs) -> np.ndarra
|
||||
|
||||
return embed_text
|
||||
|
||||
|
||||
if __name__ == "__main__":
|
||||
import asyncio
|
||||
|
||||
async def main():
|
||||
result = await gpt_4o_mini_complete('How are you?')
|
||||
result = await gpt_4o_mini_complete("How are you?")
|
||||
print(result)
|
||||
|
||||
asyncio.run(main())
|
||||
|
||||
@@ -25,6 +25,7 @@ from .base import (
|
||||
)
|
||||
from .prompt import GRAPH_FIELD_SEP, PROMPTS
|
||||
|
||||
|
||||
def chunking_by_token_size(
|
||||
content: str, overlap_token_size=128, max_token_size=1024, tiktoken_model="gpt-4o"
|
||||
):
|
||||
@@ -45,6 +46,7 @@ def chunking_by_token_size(
|
||||
)
|
||||
return results
|
||||
|
||||
|
||||
async def _handle_entity_relation_summary(
|
||||
entity_or_relation_name: str,
|
||||
description: str,
|
||||
@@ -229,9 +231,10 @@ async def _merge_edges_then_upsert(
|
||||
description=description,
|
||||
keywords=keywords,
|
||||
)
|
||||
|
||||
|
||||
return edge_data
|
||||
|
||||
|
||||
async def extract_entities(
|
||||
chunks: dict[str, TextChunkSchema],
|
||||
knwoledge_graph_inst: BaseGraphStorage,
|
||||
@@ -352,7 +355,9 @@ async def extract_entities(
|
||||
logger.warning("Didn't extract any entities, maybe your LLM is not working")
|
||||
return None
|
||||
if not len(all_relationships_data):
|
||||
logger.warning("Didn't extract any relationships, maybe your LLM is not working")
|
||||
logger.warning(
|
||||
"Didn't extract any relationships, maybe your LLM is not working"
|
||||
)
|
||||
return None
|
||||
|
||||
if entity_vdb is not None:
|
||||
@@ -370,7 +375,10 @@ async def extract_entities(
|
||||
compute_mdhash_id(dp["src_id"] + dp["tgt_id"], prefix="rel-"): {
|
||||
"src_id": dp["src_id"],
|
||||
"tgt_id": dp["tgt_id"],
|
||||
"content": dp["keywords"] + dp["src_id"] + dp["tgt_id"] + dp["description"],
|
||||
"content": dp["keywords"]
|
||||
+ dp["src_id"]
|
||||
+ dp["tgt_id"]
|
||||
+ dp["description"],
|
||||
}
|
||||
for dp in all_relationships_data
|
||||
}
|
||||
@@ -378,6 +386,7 @@ async def extract_entities(
|
||||
|
||||
return knwoledge_graph_inst
|
||||
|
||||
|
||||
async def local_query(
|
||||
query,
|
||||
knowledge_graph_inst: BaseGraphStorage,
|
||||
@@ -393,19 +402,24 @@ async def local_query(
|
||||
kw_prompt_temp = PROMPTS["keywords_extraction"]
|
||||
kw_prompt = kw_prompt_temp.format(query=query)
|
||||
result = await use_model_func(kw_prompt)
|
||||
|
||||
|
||||
try:
|
||||
keywords_data = json.loads(result)
|
||||
keywords = keywords_data.get("low_level_keywords", [])
|
||||
keywords = ', '.join(keywords)
|
||||
except json.JSONDecodeError as e:
|
||||
keywords = ", ".join(keywords)
|
||||
except json.JSONDecodeError:
|
||||
try:
|
||||
result = result.replace(kw_prompt[:-1],'').replace('user','').replace('model','').strip()
|
||||
result = '{' + result.split('{')[1].split('}')[0] + '}'
|
||||
result = (
|
||||
result.replace(kw_prompt[:-1], "")
|
||||
.replace("user", "")
|
||||
.replace("model", "")
|
||||
.strip()
|
||||
)
|
||||
result = "{" + result.split("{")[1].split("}")[0] + "}"
|
||||
|
||||
keywords_data = json.loads(result)
|
||||
keywords = keywords_data.get("low_level_keywords", [])
|
||||
keywords = ', '.join(keywords)
|
||||
keywords = ", ".join(keywords)
|
||||
# Handle parsing error
|
||||
except json.JSONDecodeError as e:
|
||||
print(f"JSON parsing error: {e}")
|
||||
@@ -430,11 +444,20 @@ async def local_query(
|
||||
query,
|
||||
system_prompt=sys_prompt,
|
||||
)
|
||||
if len(response)>len(sys_prompt):
|
||||
response = response.replace(sys_prompt,'').replace('user','').replace('model','').replace(query,'').replace('<system>','').replace('</system>','').strip()
|
||||
|
||||
if len(response) > len(sys_prompt):
|
||||
response = (
|
||||
response.replace(sys_prompt, "")
|
||||
.replace("user", "")
|
||||
.replace("model", "")
|
||||
.replace(query, "")
|
||||
.replace("<system>", "")
|
||||
.replace("</system>", "")
|
||||
.strip()
|
||||
)
|
||||
|
||||
return response
|
||||
|
||||
|
||||
async def _build_local_query_context(
|
||||
query,
|
||||
knowledge_graph_inst: BaseGraphStorage,
|
||||
@@ -516,6 +539,7 @@ async def _build_local_query_context(
|
||||
```
|
||||
"""
|
||||
|
||||
|
||||
async def _find_most_related_text_unit_from_entities(
|
||||
node_datas: list[dict],
|
||||
query_param: QueryParam,
|
||||
@@ -576,6 +600,7 @@ async def _find_most_related_text_unit_from_entities(
|
||||
all_text_units: list[TextChunkSchema] = [t["data"] for t in all_text_units]
|
||||
return all_text_units
|
||||
|
||||
|
||||
async def _find_most_related_edges_from_entities(
|
||||
node_datas: list[dict],
|
||||
query_param: QueryParam,
|
||||
@@ -609,6 +634,7 @@ async def _find_most_related_edges_from_entities(
|
||||
)
|
||||
return all_edges_data
|
||||
|
||||
|
||||
async def global_query(
|
||||
query,
|
||||
knowledge_graph_inst: BaseGraphStorage,
|
||||
@@ -624,20 +650,25 @@ async def global_query(
|
||||
kw_prompt_temp = PROMPTS["keywords_extraction"]
|
||||
kw_prompt = kw_prompt_temp.format(query=query)
|
||||
result = await use_model_func(kw_prompt)
|
||||
|
||||
|
||||
try:
|
||||
keywords_data = json.loads(result)
|
||||
keywords = keywords_data.get("high_level_keywords", [])
|
||||
keywords = ', '.join(keywords)
|
||||
except json.JSONDecodeError as e:
|
||||
keywords = ", ".join(keywords)
|
||||
except json.JSONDecodeError:
|
||||
try:
|
||||
result = result.replace(kw_prompt[:-1],'').replace('user','').replace('model','').strip()
|
||||
result = '{' + result.split('{')[1].split('}')[0] + '}'
|
||||
result = (
|
||||
result.replace(kw_prompt[:-1], "")
|
||||
.replace("user", "")
|
||||
.replace("model", "")
|
||||
.strip()
|
||||
)
|
||||
result = "{" + result.split("{")[1].split("}")[0] + "}"
|
||||
|
||||
keywords_data = json.loads(result)
|
||||
keywords = keywords_data.get("high_level_keywords", [])
|
||||
keywords = ', '.join(keywords)
|
||||
|
||||
keywords = ", ".join(keywords)
|
||||
|
||||
except json.JSONDecodeError as e:
|
||||
# Handle parsing error
|
||||
print(f"JSON parsing error: {e}")
|
||||
@@ -651,12 +682,12 @@ async def global_query(
|
||||
text_chunks_db,
|
||||
query_param,
|
||||
)
|
||||
|
||||
|
||||
if query_param.only_need_context:
|
||||
return context
|
||||
if context is None:
|
||||
return PROMPTS["fail_response"]
|
||||
|
||||
|
||||
sys_prompt_temp = PROMPTS["rag_response"]
|
||||
sys_prompt = sys_prompt_temp.format(
|
||||
context_data=context, response_type=query_param.response_type
|
||||
@@ -665,11 +696,20 @@ async def global_query(
|
||||
query,
|
||||
system_prompt=sys_prompt,
|
||||
)
|
||||
if len(response)>len(sys_prompt):
|
||||
response = response.replace(sys_prompt,'').replace('user','').replace('model','').replace(query,'').replace('<system>','').replace('</system>','').strip()
|
||||
|
||||
if len(response) > len(sys_prompt):
|
||||
response = (
|
||||
response.replace(sys_prompt, "")
|
||||
.replace("user", "")
|
||||
.replace("model", "")
|
||||
.replace(query, "")
|
||||
.replace("<system>", "")
|
||||
.replace("</system>", "")
|
||||
.strip()
|
||||
)
|
||||
|
||||
return response
|
||||
|
||||
|
||||
async def _build_global_query_context(
|
||||
keywords,
|
||||
knowledge_graph_inst: BaseGraphStorage,
|
||||
@@ -679,14 +719,14 @@ async def _build_global_query_context(
|
||||
query_param: QueryParam,
|
||||
):
|
||||
results = await relationships_vdb.query(keywords, top_k=query_param.top_k)
|
||||
|
||||
|
||||
if not len(results):
|
||||
return None
|
||||
|
||||
|
||||
edge_datas = await asyncio.gather(
|
||||
*[knowledge_graph_inst.get_edge(r["src_id"], r["tgt_id"]) for r in results]
|
||||
)
|
||||
|
||||
|
||||
if not all([n is not None for n in edge_datas]):
|
||||
logger.warning("Some edges are missing, maybe the storage is damaged")
|
||||
edge_degree = await asyncio.gather(
|
||||
@@ -765,6 +805,7 @@ async def _build_global_query_context(
|
||||
```
|
||||
"""
|
||||
|
||||
|
||||
async def _find_most_related_entities_from_relationships(
|
||||
edge_datas: list[dict],
|
||||
query_param: QueryParam,
|
||||
@@ -774,7 +815,7 @@ async def _find_most_related_entities_from_relationships(
|
||||
for e in edge_datas:
|
||||
entity_names.add(e["src_id"])
|
||||
entity_names.add(e["tgt_id"])
|
||||
|
||||
|
||||
node_datas = await asyncio.gather(
|
||||
*[knowledge_graph_inst.get_node(entity_name) for entity_name in entity_names]
|
||||
)
|
||||
@@ -795,13 +836,13 @@ async def _find_most_related_entities_from_relationships(
|
||||
|
||||
return node_datas
|
||||
|
||||
|
||||
async def _find_related_text_unit_from_relationships(
|
||||
edge_datas: list[dict],
|
||||
query_param: QueryParam,
|
||||
text_chunks_db: BaseKVStorage[TextChunkSchema],
|
||||
knowledge_graph_inst: BaseGraphStorage,
|
||||
):
|
||||
|
||||
text_units = [
|
||||
split_string_by_multi_markers(dp["source_id"], [GRAPH_FIELD_SEP])
|
||||
for dp in edge_datas
|
||||
@@ -816,15 +857,13 @@ async def _find_related_text_unit_from_relationships(
|
||||
"data": await text_chunks_db.get_by_id(c_id),
|
||||
"order": index,
|
||||
}
|
||||
|
||||
|
||||
if any([v is None for v in all_text_units_lookup.values()]):
|
||||
logger.warning("Text chunks are missing, maybe the storage is damaged")
|
||||
all_text_units = [
|
||||
{"id": k, **v} for k, v in all_text_units_lookup.items() if v is not None
|
||||
]
|
||||
all_text_units = sorted(
|
||||
all_text_units, key=lambda x: x["order"]
|
||||
)
|
||||
all_text_units = sorted(all_text_units, key=lambda x: x["order"])
|
||||
all_text_units = truncate_list_by_token_size(
|
||||
all_text_units,
|
||||
key=lambda x: x["data"]["content"],
|
||||
@@ -834,6 +873,7 @@ async def _find_related_text_unit_from_relationships(
|
||||
|
||||
return all_text_units
|
||||
|
||||
|
||||
async def hybrid_query(
|
||||
query,
|
||||
knowledge_graph_inst: BaseGraphStorage,
|
||||
@@ -849,24 +889,29 @@ async def hybrid_query(
|
||||
|
||||
kw_prompt_temp = PROMPTS["keywords_extraction"]
|
||||
kw_prompt = kw_prompt_temp.format(query=query)
|
||||
|
||||
|
||||
result = await use_model_func(kw_prompt)
|
||||
try:
|
||||
keywords_data = json.loads(result)
|
||||
hl_keywords = keywords_data.get("high_level_keywords", [])
|
||||
ll_keywords = keywords_data.get("low_level_keywords", [])
|
||||
hl_keywords = ', '.join(hl_keywords)
|
||||
ll_keywords = ', '.join(ll_keywords)
|
||||
except json.JSONDecodeError as e:
|
||||
hl_keywords = ", ".join(hl_keywords)
|
||||
ll_keywords = ", ".join(ll_keywords)
|
||||
except json.JSONDecodeError:
|
||||
try:
|
||||
result = result.replace(kw_prompt[:-1],'').replace('user','').replace('model','').strip()
|
||||
result = '{' + result.split('{')[1].split('}')[0] + '}'
|
||||
result = (
|
||||
result.replace(kw_prompt[:-1], "")
|
||||
.replace("user", "")
|
||||
.replace("model", "")
|
||||
.strip()
|
||||
)
|
||||
result = "{" + result.split("{")[1].split("}")[0] + "}"
|
||||
|
||||
keywords_data = json.loads(result)
|
||||
hl_keywords = keywords_data.get("high_level_keywords", [])
|
||||
ll_keywords = keywords_data.get("low_level_keywords", [])
|
||||
hl_keywords = ', '.join(hl_keywords)
|
||||
ll_keywords = ', '.join(ll_keywords)
|
||||
hl_keywords = ", ".join(hl_keywords)
|
||||
ll_keywords = ", ".join(ll_keywords)
|
||||
# Handle parsing error
|
||||
except json.JSONDecodeError as e:
|
||||
print(f"JSON parsing error: {e}")
|
||||
@@ -897,7 +942,7 @@ async def hybrid_query(
|
||||
return context
|
||||
if context is None:
|
||||
return PROMPTS["fail_response"]
|
||||
|
||||
|
||||
sys_prompt_temp = PROMPTS["rag_response"]
|
||||
sys_prompt = sys_prompt_temp.format(
|
||||
context_data=context, response_type=query_param.response_type
|
||||
@@ -906,53 +951,78 @@ async def hybrid_query(
|
||||
query,
|
||||
system_prompt=sys_prompt,
|
||||
)
|
||||
if len(response)>len(sys_prompt):
|
||||
response = response.replace(sys_prompt,'').replace('user','').replace('model','').replace(query,'').replace('<system>','').replace('</system>','').strip()
|
||||
if len(response) > len(sys_prompt):
|
||||
response = (
|
||||
response.replace(sys_prompt, "")
|
||||
.replace("user", "")
|
||||
.replace("model", "")
|
||||
.replace(query, "")
|
||||
.replace("<system>", "")
|
||||
.replace("</system>", "")
|
||||
.strip()
|
||||
)
|
||||
return response
|
||||
|
||||
|
||||
def combine_contexts(high_level_context, low_level_context):
|
||||
# Function to extract entities, relationships, and sources from context strings
|
||||
|
||||
def extract_sections(context):
|
||||
entities_match = re.search(r'-----Entities-----\s*```csv\s*(.*?)\s*```', context, re.DOTALL)
|
||||
relationships_match = re.search(r'-----Relationships-----\s*```csv\s*(.*?)\s*```', context, re.DOTALL)
|
||||
sources_match = re.search(r'-----Sources-----\s*```csv\s*(.*?)\s*```', context, re.DOTALL)
|
||||
|
||||
entities = entities_match.group(1) if entities_match else ''
|
||||
relationships = relationships_match.group(1) if relationships_match else ''
|
||||
sources = sources_match.group(1) if sources_match else ''
|
||||
|
||||
entities_match = re.search(
|
||||
r"-----Entities-----\s*```csv\s*(.*?)\s*```", context, re.DOTALL
|
||||
)
|
||||
relationships_match = re.search(
|
||||
r"-----Relationships-----\s*```csv\s*(.*?)\s*```", context, re.DOTALL
|
||||
)
|
||||
sources_match = re.search(
|
||||
r"-----Sources-----\s*```csv\s*(.*?)\s*```", context, re.DOTALL
|
||||
)
|
||||
|
||||
entities = entities_match.group(1) if entities_match else ""
|
||||
relationships = relationships_match.group(1) if relationships_match else ""
|
||||
sources = sources_match.group(1) if sources_match else ""
|
||||
|
||||
return entities, relationships, sources
|
||||
|
||||
|
||||
# Extract sections from both contexts
|
||||
|
||||
if high_level_context==None:
|
||||
warnings.warn("High Level context is None. Return empty High entity/relationship/source")
|
||||
hl_entities, hl_relationships, hl_sources = '','',''
|
||||
if high_level_context is None:
|
||||
warnings.warn(
|
||||
"High Level context is None. Return empty High entity/relationship/source"
|
||||
)
|
||||
hl_entities, hl_relationships, hl_sources = "", "", ""
|
||||
else:
|
||||
hl_entities, hl_relationships, hl_sources = extract_sections(high_level_context)
|
||||
|
||||
|
||||
if low_level_context==None:
|
||||
warnings.warn("Low Level context is None. Return empty Low entity/relationship/source")
|
||||
ll_entities, ll_relationships, ll_sources = '','',''
|
||||
if low_level_context is None:
|
||||
warnings.warn(
|
||||
"Low Level context is None. Return empty Low entity/relationship/source"
|
||||
)
|
||||
ll_entities, ll_relationships, ll_sources = "", "", ""
|
||||
else:
|
||||
ll_entities, ll_relationships, ll_sources = extract_sections(low_level_context)
|
||||
|
||||
|
||||
|
||||
# Combine and deduplicate the entities
|
||||
combined_entities_set = set(filter(None, hl_entities.strip().split('\n') + ll_entities.strip().split('\n')))
|
||||
combined_entities = '\n'.join(combined_entities_set)
|
||||
|
||||
combined_entities_set = set(
|
||||
filter(None, hl_entities.strip().split("\n") + ll_entities.strip().split("\n"))
|
||||
)
|
||||
combined_entities = "\n".join(combined_entities_set)
|
||||
|
||||
# Combine and deduplicate the relationships
|
||||
combined_relationships_set = set(filter(None, hl_relationships.strip().split('\n') + ll_relationships.strip().split('\n')))
|
||||
combined_relationships = '\n'.join(combined_relationships_set)
|
||||
|
||||
combined_relationships_set = set(
|
||||
filter(
|
||||
None,
|
||||
hl_relationships.strip().split("\n") + ll_relationships.strip().split("\n"),
|
||||
)
|
||||
)
|
||||
combined_relationships = "\n".join(combined_relationships_set)
|
||||
|
||||
# Combine and deduplicate the sources
|
||||
combined_sources_set = set(filter(None, hl_sources.strip().split('\n') + ll_sources.strip().split('\n')))
|
||||
combined_sources = '\n'.join(combined_sources_set)
|
||||
|
||||
combined_sources_set = set(
|
||||
filter(None, hl_sources.strip().split("\n") + ll_sources.strip().split("\n"))
|
||||
)
|
||||
combined_sources = "\n".join(combined_sources_set)
|
||||
|
||||
# Format the combined context
|
||||
return f"""
|
||||
-----Entities-----
|
||||
@@ -964,6 +1034,7 @@ def combine_contexts(high_level_context, low_level_context):
|
||||
{combined_sources}
|
||||
"""
|
||||
|
||||
|
||||
async def naive_query(
|
||||
query,
|
||||
chunks_vdb: BaseVectorStorage,
|
||||
@@ -996,8 +1067,16 @@ async def naive_query(
|
||||
system_prompt=sys_prompt,
|
||||
)
|
||||
|
||||
if len(response)>len(sys_prompt):
|
||||
response = response[len(sys_prompt):].replace(sys_prompt,'').replace('user','').replace('model','').replace(query,'').replace('<system>','').replace('</system>','').strip()
|
||||
|
||||
return response
|
||||
if len(response) > len(sys_prompt):
|
||||
response = (
|
||||
response[len(sys_prompt) :]
|
||||
.replace(sys_prompt, "")
|
||||
.replace("user", "")
|
||||
.replace("model", "")
|
||||
.replace(query, "")
|
||||
.replace("<system>", "")
|
||||
.replace("</system>", "")
|
||||
.strip()
|
||||
)
|
||||
|
||||
return response
|
||||
|
||||
@@ -9,9 +9,7 @@ PROMPTS["process_tickers"] = ["⠋", "⠙", "⠹", "⠸", "⠼", "⠴", "⠦", "
|
||||
|
||||
PROMPTS["DEFAULT_ENTITY_TYPES"] = ["organization", "person", "geo", "event"]
|
||||
|
||||
PROMPTS[
|
||||
"entity_extraction"
|
||||
] = """-Goal-
|
||||
PROMPTS["entity_extraction"] = """-Goal-
|
||||
Given a text document that is potentially relevant to this activity and a list of entity types, identify all entities of those types from the text and all relationships among the identified entities.
|
||||
|
||||
-Steps-
|
||||
@@ -32,7 +30,7 @@ Format each relationship as ("relationship"{tuple_delimiter}<source_entity>{tupl
|
||||
|
||||
3. Identify high-level key words that summarize the main concepts, themes, or topics of the entire text. These should capture the overarching ideas present in the document.
|
||||
Format the content-level key words as ("content_keywords"{tuple_delimiter}<high_level_keywords>)
|
||||
|
||||
|
||||
4. Return output in English as a single list of all the entities and relationships identified in steps 1 and 2. Use **{record_delimiter}** as the list delimiter.
|
||||
|
||||
5. When finished, output {completion_delimiter}
|
||||
@@ -146,9 +144,7 @@ PROMPTS[
|
||||
|
||||
PROMPTS["fail_response"] = "Sorry, I'm not able to provide an answer to that question."
|
||||
|
||||
PROMPTS[
|
||||
"rag_response"
|
||||
] = """---Role---
|
||||
PROMPTS["rag_response"] = """---Role---
|
||||
|
||||
You are a helpful assistant responding to questions about data in the tables provided.
|
||||
|
||||
@@ -163,25 +159,10 @@ Do not include information where the supporting evidence for it is not provided.
|
||||
|
||||
{response_type}
|
||||
|
||||
|
||||
---Data tables---
|
||||
|
||||
{context_data}
|
||||
|
||||
|
||||
---Goal---
|
||||
|
||||
Generate a response of the target length and format that responds to the user's question, summarizing all information in the input data tables appropriate for the response length and format, and incorporating any relevant general knowledge.
|
||||
|
||||
If you don't know the answer, just say so. Do not make anything up.
|
||||
|
||||
Do not include information where the supporting evidence for it is not provided.
|
||||
|
||||
|
||||
---Target response length and format---
|
||||
|
||||
{response_type}
|
||||
|
||||
Add sections and commentary to the response as appropriate for the length and format. Style the response in markdown.
|
||||
"""
|
||||
|
||||
@@ -241,9 +222,7 @@ Output:
|
||||
|
||||
"""
|
||||
|
||||
PROMPTS[
|
||||
"naive_rag_response"
|
||||
] = """You're a helpful assistant
|
||||
PROMPTS["naive_rag_response"] = """You're a helpful assistant
|
||||
Below are the knowledge you know:
|
||||
{content_data}
|
||||
---
|
||||
|
||||
@@ -1,16 +1,11 @@
|
||||
import asyncio
|
||||
import html
|
||||
import json
|
||||
import os
|
||||
from collections import defaultdict
|
||||
from dataclasses import dataclass, field
|
||||
from dataclasses import dataclass
|
||||
from typing import Any, Union, cast
|
||||
import pickle
|
||||
import hnswlib
|
||||
import networkx as nx
|
||||
import numpy as np
|
||||
from nano_vectordb import NanoVectorDB
|
||||
import xxhash
|
||||
|
||||
from .utils import load_json, logger, write_json
|
||||
from .base import (
|
||||
@@ -19,6 +14,7 @@ from .base import (
|
||||
BaseVectorStorage,
|
||||
)
|
||||
|
||||
|
||||
@dataclass
|
||||
class JsonKVStorage(BaseKVStorage):
|
||||
def __post_init__(self):
|
||||
@@ -59,12 +55,12 @@ class JsonKVStorage(BaseKVStorage):
|
||||
async def drop(self):
|
||||
self._data = {}
|
||||
|
||||
|
||||
@dataclass
|
||||
class NanoVectorDBStorage(BaseVectorStorage):
|
||||
cosine_better_than_threshold: float = 0.2
|
||||
|
||||
def __post_init__(self):
|
||||
|
||||
self._client_file_name = os.path.join(
|
||||
self.global_config["working_dir"], f"vdb_{self.namespace}.json"
|
||||
)
|
||||
@@ -118,6 +114,7 @@ class NanoVectorDBStorage(BaseVectorStorage):
|
||||
async def index_done_callback(self):
|
||||
self._client.save()
|
||||
|
||||
|
||||
@dataclass
|
||||
class NetworkXStorage(BaseGraphStorage):
|
||||
@staticmethod
|
||||
@@ -142,7 +139,9 @@ class NetworkXStorage(BaseGraphStorage):
|
||||
|
||||
graph = graph.copy()
|
||||
graph = cast(nx.Graph, largest_connected_component(graph))
|
||||
node_mapping = {node: html.unescape(node.upper().strip()) for node in graph.nodes()} # type: ignore
|
||||
node_mapping = {
|
||||
node: html.unescape(node.upper().strip()) for node in graph.nodes()
|
||||
} # type: ignore
|
||||
graph = nx.relabel_nodes(graph, node_mapping)
|
||||
return NetworkXStorage._stabilize_graph(graph)
|
||||
|
||||
|
||||
@@ -8,6 +8,7 @@ from dataclasses import dataclass
|
||||
from functools import wraps
|
||||
from hashlib import md5
|
||||
from typing import Any, Union
|
||||
import xml.etree.ElementTree as ET
|
||||
|
||||
import numpy as np
|
||||
import tiktoken
|
||||
@@ -16,18 +17,22 @@ ENCODER = None
|
||||
|
||||
logger = logging.getLogger("lightrag")
|
||||
|
||||
|
||||
def set_logger(log_file: str):
|
||||
logger.setLevel(logging.DEBUG)
|
||||
|
||||
file_handler = logging.FileHandler(log_file)
|
||||
file_handler.setLevel(logging.DEBUG)
|
||||
|
||||
formatter = logging.Formatter('%(asctime)s - %(name)s - %(levelname)s - %(message)s')
|
||||
formatter = logging.Formatter(
|
||||
"%(asctime)s - %(name)s - %(levelname)s - %(message)s"
|
||||
)
|
||||
file_handler.setFormatter(formatter)
|
||||
|
||||
if not logger.handlers:
|
||||
logger.addHandler(file_handler)
|
||||
|
||||
|
||||
@dataclass
|
||||
class EmbeddingFunc:
|
||||
embedding_dim: int
|
||||
@@ -36,7 +41,8 @@ class EmbeddingFunc:
|
||||
|
||||
async def __call__(self, *args, **kwargs) -> np.ndarray:
|
||||
return await self.func(*args, **kwargs)
|
||||
|
||||
|
||||
|
||||
def locate_json_string_body_from_string(content: str) -> Union[str, None]:
|
||||
"""Locate the JSON string body from a string"""
|
||||
maybe_json_str = re.search(r"{.*}", content, re.DOTALL)
|
||||
@@ -45,6 +51,7 @@ def locate_json_string_body_from_string(content: str) -> Union[str, None]:
|
||||
else:
|
||||
return None
|
||||
|
||||
|
||||
def convert_response_to_json(response: str) -> dict:
|
||||
json_str = locate_json_string_body_from_string(response)
|
||||
assert json_str is not None, f"Unable to parse JSON from response: {response}"
|
||||
@@ -55,12 +62,15 @@ def convert_response_to_json(response: str) -> dict:
|
||||
logger.error(f"Failed to parse JSON: {json_str}")
|
||||
raise e from None
|
||||
|
||||
|
||||
def compute_args_hash(*args):
|
||||
return md5(str(args).encode()).hexdigest()
|
||||
|
||||
|
||||
def compute_mdhash_id(content, prefix: str = ""):
|
||||
return prefix + md5(content.encode()).hexdigest()
|
||||
|
||||
|
||||
def limit_async_func_call(max_size: int, waitting_time: float = 0.0001):
|
||||
"""Add restriction of maximum async calling times for a async func"""
|
||||
|
||||
@@ -82,6 +92,7 @@ def limit_async_func_call(max_size: int, waitting_time: float = 0.0001):
|
||||
|
||||
return final_decro
|
||||
|
||||
|
||||
def wrap_embedding_func_with_attrs(**kwargs):
|
||||
"""Wrap a function with attributes"""
|
||||
|
||||
@@ -91,16 +102,19 @@ def wrap_embedding_func_with_attrs(**kwargs):
|
||||
|
||||
return final_decro
|
||||
|
||||
|
||||
def load_json(file_name):
|
||||
if not os.path.exists(file_name):
|
||||
return None
|
||||
with open(file_name, encoding="utf-8") as f:
|
||||
return json.load(f)
|
||||
|
||||
|
||||
def write_json(json_obj, file_name):
|
||||
with open(file_name, "w", encoding="utf-8") as f:
|
||||
json.dump(json_obj, f, indent=2, ensure_ascii=False)
|
||||
|
||||
|
||||
def encode_string_by_tiktoken(content: str, model_name: str = "gpt-4o"):
|
||||
global ENCODER
|
||||
if ENCODER is None:
|
||||
@@ -116,12 +130,14 @@ def decode_tokens_by_tiktoken(tokens: list[int], model_name: str = "gpt-4o"):
|
||||
content = ENCODER.decode(tokens)
|
||||
return content
|
||||
|
||||
|
||||
def pack_user_ass_to_openai_messages(*args: str):
|
||||
roles = ["user", "assistant"]
|
||||
return [
|
||||
{"role": roles[i % 2], "content": content} for i, content in enumerate(args)
|
||||
]
|
||||
|
||||
|
||||
def split_string_by_multi_markers(content: str, markers: list[str]) -> list[str]:
|
||||
"""Split a string by multiple markers"""
|
||||
if not markers:
|
||||
@@ -129,6 +145,7 @@ def split_string_by_multi_markers(content: str, markers: list[str]) -> list[str]
|
||||
results = re.split("|".join(re.escape(marker) for marker in markers), content)
|
||||
return [r.strip() for r in results if r.strip()]
|
||||
|
||||
|
||||
# Refer the utils functions of the official GraphRAG implementation:
|
||||
# https://github.com/microsoft/graphrag
|
||||
def clean_str(input: Any) -> str:
|
||||
@@ -141,9 +158,11 @@ def clean_str(input: Any) -> str:
|
||||
# https://stackoverflow.com/questions/4324790/removing-control-characters-from-a-string-in-python
|
||||
return re.sub(r"[\x00-\x1f\x7f-\x9f]", "", result)
|
||||
|
||||
|
||||
def is_float_regex(value):
|
||||
return bool(re.match(r"^[-+]?[0-9]*\.?[0-9]+$", value))
|
||||
|
||||
|
||||
def truncate_list_by_token_size(list_data: list, key: callable, max_token_size: int):
|
||||
"""Truncate a list of data by token size"""
|
||||
if max_token_size <= 0:
|
||||
@@ -155,11 +174,61 @@ def truncate_list_by_token_size(list_data: list, key: callable, max_token_size:
|
||||
return list_data[:i]
|
||||
return list_data
|
||||
|
||||
|
||||
def list_of_list_to_csv(data: list[list]):
|
||||
return "\n".join(
|
||||
[",\t".join([str(data_dd) for data_dd in data_d]) for data_d in data]
|
||||
)
|
||||
|
||||
|
||||
def save_data_to_file(data, file_name):
|
||||
with open(file_name, 'w', encoding='utf-8') as f:
|
||||
json.dump(data, f, ensure_ascii=False, indent=4)
|
||||
with open(file_name, "w", encoding="utf-8") as f:
|
||||
json.dump(data, f, ensure_ascii=False, indent=4)
|
||||
|
||||
def xml_to_json(xml_file):
|
||||
try:
|
||||
tree = ET.parse(xml_file)
|
||||
root = tree.getroot()
|
||||
|
||||
# Print the root element's tag and attributes to confirm the file has been correctly loaded
|
||||
print(f"Root element: {root.tag}")
|
||||
print(f"Root attributes: {root.attrib}")
|
||||
|
||||
data = {
|
||||
"nodes": [],
|
||||
"edges": []
|
||||
}
|
||||
|
||||
# Use namespace
|
||||
namespace = {'': 'http://graphml.graphdrawing.org/xmlns'}
|
||||
|
||||
for node in root.findall('.//node', namespace):
|
||||
node_data = {
|
||||
"id": node.get('id').strip('"'),
|
||||
"entity_type": node.find("./data[@key='d0']", namespace).text.strip('"') if node.find("./data[@key='d0']", namespace) is not None else "",
|
||||
"description": node.find("./data[@key='d1']", namespace).text if node.find("./data[@key='d1']", namespace) is not None else "",
|
||||
"source_id": node.find("./data[@key='d2']", namespace).text if node.find("./data[@key='d2']", namespace) is not None else ""
|
||||
}
|
||||
data["nodes"].append(node_data)
|
||||
|
||||
for edge in root.findall('.//edge', namespace):
|
||||
edge_data = {
|
||||
"source": edge.get('source').strip('"'),
|
||||
"target": edge.get('target').strip('"'),
|
||||
"weight": float(edge.find("./data[@key='d3']", namespace).text) if edge.find("./data[@key='d3']", namespace) is not None else 0.0,
|
||||
"description": edge.find("./data[@key='d4']", namespace).text if edge.find("./data[@key='d4']", namespace) is not None else "",
|
||||
"keywords": edge.find("./data[@key='d5']", namespace).text if edge.find("./data[@key='d5']", namespace) is not None else "",
|
||||
"source_id": edge.find("./data[@key='d6']", namespace).text if edge.find("./data[@key='d6']", namespace) is not None else ""
|
||||
}
|
||||
data["edges"].append(edge_data)
|
||||
|
||||
# Print the number of nodes and edges found
|
||||
print(f"Found {len(data['nodes'])} nodes and {len(data['edges'])} edges")
|
||||
|
||||
return data
|
||||
except ET.ParseError as e:
|
||||
print(f"Error parsing XML file: {e}")
|
||||
return None
|
||||
except Exception as e:
|
||||
print(f"An error occurred: {e}")
|
||||
return None
|
||||
|
||||
@@ -3,11 +3,11 @@ import json
|
||||
import glob
|
||||
import argparse
|
||||
|
||||
def extract_unique_contexts(input_directory, output_directory):
|
||||
|
||||
def extract_unique_contexts(input_directory, output_directory):
|
||||
os.makedirs(output_directory, exist_ok=True)
|
||||
|
||||
jsonl_files = glob.glob(os.path.join(input_directory, '*.jsonl'))
|
||||
jsonl_files = glob.glob(os.path.join(input_directory, "*.jsonl"))
|
||||
print(f"Found {len(jsonl_files)} JSONL files.")
|
||||
|
||||
for file_path in jsonl_files:
|
||||
@@ -21,18 +21,20 @@ def extract_unique_contexts(input_directory, output_directory):
|
||||
print(f"Processing file: {filename}")
|
||||
|
||||
try:
|
||||
with open(file_path, 'r', encoding='utf-8') as infile:
|
||||
with open(file_path, "r", encoding="utf-8") as infile:
|
||||
for line_number, line in enumerate(infile, start=1):
|
||||
line = line.strip()
|
||||
if not line:
|
||||
continue
|
||||
try:
|
||||
json_obj = json.loads(line)
|
||||
context = json_obj.get('context')
|
||||
context = json_obj.get("context")
|
||||
if context and context not in unique_contexts_dict:
|
||||
unique_contexts_dict[context] = None
|
||||
except json.JSONDecodeError as e:
|
||||
print(f"JSON decoding error in file {filename} at line {line_number}: {e}")
|
||||
print(
|
||||
f"JSON decoding error in file {filename} at line {line_number}: {e}"
|
||||
)
|
||||
except FileNotFoundError:
|
||||
print(f"File not found: {filename}")
|
||||
continue
|
||||
@@ -41,10 +43,12 @@ def extract_unique_contexts(input_directory, output_directory):
|
||||
continue
|
||||
|
||||
unique_contexts_list = list(unique_contexts_dict.keys())
|
||||
print(f"There are {len(unique_contexts_list)} unique `context` entries in the file {filename}.")
|
||||
print(
|
||||
f"There are {len(unique_contexts_list)} unique `context` entries in the file {filename}."
|
||||
)
|
||||
|
||||
try:
|
||||
with open(output_path, 'w', encoding='utf-8') as outfile:
|
||||
with open(output_path, "w", encoding="utf-8") as outfile:
|
||||
json.dump(unique_contexts_list, outfile, ensure_ascii=False, indent=4)
|
||||
print(f"Unique `context` entries have been saved to: {output_filename}")
|
||||
except Exception as e:
|
||||
@@ -55,8 +59,10 @@ def extract_unique_contexts(input_directory, output_directory):
|
||||
|
||||
if __name__ == "__main__":
|
||||
parser = argparse.ArgumentParser()
|
||||
parser.add_argument('-i', '--input_dir', type=str, default='../datasets')
|
||||
parser.add_argument('-o', '--output_dir', type=str, default='../datasets/unique_contexts')
|
||||
parser.add_argument("-i", "--input_dir", type=str, default="../datasets")
|
||||
parser.add_argument(
|
||||
"-o", "--output_dir", type=str, default="../datasets/unique_contexts"
|
||||
)
|
||||
|
||||
args = parser.parse_args()
|
||||
|
||||
|
||||
@@ -4,10 +4,11 @@ import time
|
||||
|
||||
from lightrag import LightRAG
|
||||
|
||||
|
||||
def insert_text(rag, file_path):
|
||||
with open(file_path, mode='r') as f:
|
||||
with open(file_path, mode="r") as f:
|
||||
unique_contexts = json.load(f)
|
||||
|
||||
|
||||
retries = 0
|
||||
max_retries = 3
|
||||
while retries < max_retries:
|
||||
@@ -21,6 +22,7 @@ def insert_text(rag, file_path):
|
||||
if retries == max_retries:
|
||||
print("Insertion failed after exceeding the maximum number of retries")
|
||||
|
||||
|
||||
cls = "agriculture"
|
||||
WORKING_DIR = "../{cls}"
|
||||
|
||||
@@ -29,4 +31,4 @@ if not os.path.exists(WORKING_DIR):
|
||||
|
||||
rag = LightRAG(working_dir=WORKING_DIR)
|
||||
|
||||
insert_text(rag, f"../datasets/unique_contexts/{cls}_unique_contexts.json")
|
||||
insert_text(rag, f"../datasets/unique_contexts/{cls}_unique_contexts.json")
|
||||
|
||||
@@ -7,6 +7,7 @@ from lightrag import LightRAG
|
||||
from lightrag.utils import EmbeddingFunc
|
||||
from lightrag.llm import openai_complete_if_cache, openai_embedding
|
||||
|
||||
|
||||
## For Upstage API
|
||||
# please check if embedding_dim=4096 in lightrag.py and llm.py in lightrag direcotry
|
||||
async def llm_model_func(
|
||||
@@ -19,22 +20,26 @@ async def llm_model_func(
|
||||
history_messages=history_messages,
|
||||
api_key=os.getenv("UPSTAGE_API_KEY"),
|
||||
base_url="https://api.upstage.ai/v1/solar",
|
||||
**kwargs
|
||||
**kwargs,
|
||||
)
|
||||
|
||||
|
||||
async def embedding_func(texts: list[str]) -> np.ndarray:
|
||||
return await openai_embedding(
|
||||
texts,
|
||||
model="solar-embedding-1-large-query",
|
||||
api_key=os.getenv("UPSTAGE_API_KEY"),
|
||||
base_url="https://api.upstage.ai/v1/solar"
|
||||
base_url="https://api.upstage.ai/v1/solar",
|
||||
)
|
||||
|
||||
|
||||
## /For Upstage API
|
||||
|
||||
|
||||
def insert_text(rag, file_path):
|
||||
with open(file_path, mode='r') as f:
|
||||
with open(file_path, mode="r") as f:
|
||||
unique_contexts = json.load(f)
|
||||
|
||||
|
||||
retries = 0
|
||||
max_retries = 3
|
||||
while retries < max_retries:
|
||||
@@ -48,19 +53,19 @@ def insert_text(rag, file_path):
|
||||
if retries == max_retries:
|
||||
print("Insertion failed after exceeding the maximum number of retries")
|
||||
|
||||
|
||||
cls = "mix"
|
||||
WORKING_DIR = f"../{cls}"
|
||||
|
||||
if not os.path.exists(WORKING_DIR):
|
||||
os.mkdir(WORKING_DIR)
|
||||
|
||||
rag = LightRAG(working_dir=WORKING_DIR,
|
||||
llm_model_func=llm_model_func,
|
||||
embedding_func=EmbeddingFunc(
|
||||
embedding_dim=4096,
|
||||
max_token_size=8192,
|
||||
func=embedding_func
|
||||
)
|
||||
)
|
||||
rag = LightRAG(
|
||||
working_dir=WORKING_DIR,
|
||||
llm_model_func=llm_model_func,
|
||||
embedding_func=EmbeddingFunc(
|
||||
embedding_dim=4096, max_token_size=8192, func=embedding_func
|
||||
),
|
||||
)
|
||||
|
||||
insert_text(rag, f"../datasets/unique_contexts/{cls}_unique_contexts.json")
|
||||
|
||||
@@ -1,8 +1,8 @@
|
||||
import os
|
||||
import json
|
||||
from openai import OpenAI
|
||||
from transformers import GPT2Tokenizer
|
||||
|
||||
|
||||
def openai_complete_if_cache(
|
||||
model="gpt-4o", prompt=None, system_prompt=None, history_messages=[], **kwargs
|
||||
) -> str:
|
||||
@@ -19,24 +19,26 @@ def openai_complete_if_cache(
|
||||
)
|
||||
return response.choices[0].message.content
|
||||
|
||||
tokenizer = GPT2Tokenizer.from_pretrained('gpt2')
|
||||
|
||||
tokenizer = GPT2Tokenizer.from_pretrained("gpt2")
|
||||
|
||||
|
||||
def get_summary(context, tot_tokens=2000):
|
||||
tokens = tokenizer.tokenize(context)
|
||||
half_tokens = tot_tokens // 2
|
||||
|
||||
start_tokens = tokens[1000:1000 + half_tokens]
|
||||
end_tokens = tokens[-(1000 + half_tokens):1000]
|
||||
start_tokens = tokens[1000 : 1000 + half_tokens]
|
||||
end_tokens = tokens[-(1000 + half_tokens) : 1000]
|
||||
|
||||
summary_tokens = start_tokens + end_tokens
|
||||
summary = tokenizer.convert_tokens_to_string(summary_tokens)
|
||||
|
||||
|
||||
return summary
|
||||
|
||||
|
||||
clses = ['agriculture']
|
||||
clses = ["agriculture"]
|
||||
for cls in clses:
|
||||
with open(f'../datasets/unique_contexts/{cls}_unique_contexts.json', mode='r') as f:
|
||||
with open(f"../datasets/unique_contexts/{cls}_unique_contexts.json", mode="r") as f:
|
||||
unique_contexts = json.load(f)
|
||||
|
||||
summaries = [get_summary(context) for context in unique_contexts]
|
||||
@@ -67,10 +69,10 @@ for cls in clses:
|
||||
...
|
||||
"""
|
||||
|
||||
result = openai_complete_if_cache(model='gpt-4o', prompt=prompt)
|
||||
result = openai_complete_if_cache(model="gpt-4o", prompt=prompt)
|
||||
|
||||
file_path = f"../datasets/questions/{cls}_questions.txt"
|
||||
with open(file_path, "w") as file:
|
||||
file.write(result)
|
||||
|
||||
print(f"{cls}_questions written to {file_path}")
|
||||
print(f"{cls}_questions written to {file_path}")
|
||||
|
||||
@@ -4,16 +4,18 @@ import asyncio
|
||||
from lightrag import LightRAG, QueryParam
|
||||
from tqdm import tqdm
|
||||
|
||||
def extract_queries(file_path):
|
||||
with open(file_path, 'r') as f:
|
||||
data = f.read()
|
||||
|
||||
data = data.replace('**', '')
|
||||
|
||||
queries = re.findall(r'- Question \d+: (.+)', data)
|
||||
def extract_queries(file_path):
|
||||
with open(file_path, "r") as f:
|
||||
data = f.read()
|
||||
|
||||
data = data.replace("**", "")
|
||||
|
||||
queries = re.findall(r"- Question \d+: (.+)", data)
|
||||
|
||||
return queries
|
||||
|
||||
|
||||
async def process_query(query_text, rag_instance, query_param):
|
||||
try:
|
||||
result, context = await rag_instance.aquery(query_text, param=query_param)
|
||||
@@ -21,6 +23,7 @@ async def process_query(query_text, rag_instance, query_param):
|
||||
except Exception as e:
|
||||
return None, {"query": query_text, "error": str(e)}
|
||||
|
||||
|
||||
def always_get_an_event_loop() -> asyncio.AbstractEventLoop:
|
||||
try:
|
||||
loop = asyncio.get_event_loop()
|
||||
@@ -29,15 +32,22 @@ def always_get_an_event_loop() -> asyncio.AbstractEventLoop:
|
||||
asyncio.set_event_loop(loop)
|
||||
return loop
|
||||
|
||||
def run_queries_and_save_to_json(queries, rag_instance, query_param, output_file, error_file):
|
||||
|
||||
def run_queries_and_save_to_json(
|
||||
queries, rag_instance, query_param, output_file, error_file
|
||||
):
|
||||
loop = always_get_an_event_loop()
|
||||
|
||||
with open(output_file, 'a', encoding='utf-8') as result_file, open(error_file, 'a', encoding='utf-8') as err_file:
|
||||
with open(output_file, "a", encoding="utf-8") as result_file, open(
|
||||
error_file, "a", encoding="utf-8"
|
||||
) as err_file:
|
||||
result_file.write("[\n")
|
||||
first_entry = True
|
||||
|
||||
for query_text in tqdm(queries, desc="Processing queries", unit="query"):
|
||||
result, error = loop.run_until_complete(process_query(query_text, rag_instance, query_param))
|
||||
result, error = loop.run_until_complete(
|
||||
process_query(query_text, rag_instance, query_param)
|
||||
)
|
||||
|
||||
if result:
|
||||
if not first_entry:
|
||||
@@ -50,6 +60,7 @@ def run_queries_and_save_to_json(queries, rag_instance, query_param, output_file
|
||||
|
||||
result_file.write("\n]")
|
||||
|
||||
|
||||
if __name__ == "__main__":
|
||||
cls = "agriculture"
|
||||
mode = "hybrid"
|
||||
@@ -59,4 +70,6 @@ if __name__ == "__main__":
|
||||
query_param = QueryParam(mode=mode)
|
||||
|
||||
queries = extract_queries(f"../datasets/questions/{cls}_questions.txt")
|
||||
run_queries_and_save_to_json(queries, rag, query_param, f"{cls}_result.json", f"{cls}_errors.json")
|
||||
run_queries_and_save_to_json(
|
||||
queries, rag, query_param, f"{cls}_result.json", f"{cls}_errors.json"
|
||||
)
|
||||
|
||||
@@ -8,6 +8,7 @@ from lightrag.llm import openai_complete_if_cache, openai_embedding
|
||||
from lightrag.utils import EmbeddingFunc
|
||||
import numpy as np
|
||||
|
||||
|
||||
## For Upstage API
|
||||
# please check if embedding_dim=4096 in lightrag.py and llm.py in lightrag direcotry
|
||||
async def llm_model_func(
|
||||
@@ -20,28 +21,33 @@ async def llm_model_func(
|
||||
history_messages=history_messages,
|
||||
api_key=os.getenv("UPSTAGE_API_KEY"),
|
||||
base_url="https://api.upstage.ai/v1/solar",
|
||||
**kwargs
|
||||
**kwargs,
|
||||
)
|
||||
|
||||
|
||||
async def embedding_func(texts: list[str]) -> np.ndarray:
|
||||
return await openai_embedding(
|
||||
texts,
|
||||
model="solar-embedding-1-large-query",
|
||||
api_key=os.getenv("UPSTAGE_API_KEY"),
|
||||
base_url="https://api.upstage.ai/v1/solar"
|
||||
base_url="https://api.upstage.ai/v1/solar",
|
||||
)
|
||||
|
||||
|
||||
## /For Upstage API
|
||||
|
||||
def extract_queries(file_path):
|
||||
with open(file_path, 'r') as f:
|
||||
data = f.read()
|
||||
|
||||
data = data.replace('**', '')
|
||||
|
||||
queries = re.findall(r'- Question \d+: (.+)', data)
|
||||
def extract_queries(file_path):
|
||||
with open(file_path, "r") as f:
|
||||
data = f.read()
|
||||
|
||||
data = data.replace("**", "")
|
||||
|
||||
queries = re.findall(r"- Question \d+: (.+)", data)
|
||||
|
||||
return queries
|
||||
|
||||
|
||||
async def process_query(query_text, rag_instance, query_param):
|
||||
try:
|
||||
result, context = await rag_instance.aquery(query_text, param=query_param)
|
||||
@@ -49,6 +55,7 @@ async def process_query(query_text, rag_instance, query_param):
|
||||
except Exception as e:
|
||||
return None, {"query": query_text, "error": str(e)}
|
||||
|
||||
|
||||
def always_get_an_event_loop() -> asyncio.AbstractEventLoop:
|
||||
try:
|
||||
loop = asyncio.get_event_loop()
|
||||
@@ -57,15 +64,22 @@ def always_get_an_event_loop() -> asyncio.AbstractEventLoop:
|
||||
asyncio.set_event_loop(loop)
|
||||
return loop
|
||||
|
||||
def run_queries_and_save_to_json(queries, rag_instance, query_param, output_file, error_file):
|
||||
|
||||
def run_queries_and_save_to_json(
|
||||
queries, rag_instance, query_param, output_file, error_file
|
||||
):
|
||||
loop = always_get_an_event_loop()
|
||||
|
||||
with open(output_file, 'a', encoding='utf-8') as result_file, open(error_file, 'a', encoding='utf-8') as err_file:
|
||||
with open(output_file, "a", encoding="utf-8") as result_file, open(
|
||||
error_file, "a", encoding="utf-8"
|
||||
) as err_file:
|
||||
result_file.write("[\n")
|
||||
first_entry = True
|
||||
|
||||
for query_text in tqdm(queries, desc="Processing queries", unit="query"):
|
||||
result, error = loop.run_until_complete(process_query(query_text, rag_instance, query_param))
|
||||
result, error = loop.run_until_complete(
|
||||
process_query(query_text, rag_instance, query_param)
|
||||
)
|
||||
|
||||
if result:
|
||||
if not first_entry:
|
||||
@@ -78,22 +92,24 @@ def run_queries_and_save_to_json(queries, rag_instance, query_param, output_file
|
||||
|
||||
result_file.write("\n]")
|
||||
|
||||
|
||||
if __name__ == "__main__":
|
||||
cls = "mix"
|
||||
mode = "hybrid"
|
||||
WORKING_DIR = f"../{cls}"
|
||||
|
||||
rag = LightRAG(working_dir=WORKING_DIR)
|
||||
rag = LightRAG(working_dir=WORKING_DIR,
|
||||
llm_model_func=llm_model_func,
|
||||
embedding_func=EmbeddingFunc(
|
||||
embedding_dim=4096,
|
||||
max_token_size=8192,
|
||||
func=embedding_func
|
||||
)
|
||||
)
|
||||
rag = LightRAG(
|
||||
working_dir=WORKING_DIR,
|
||||
llm_model_func=llm_model_func,
|
||||
embedding_func=EmbeddingFunc(
|
||||
embedding_dim=4096, max_token_size=8192, func=embedding_func
|
||||
),
|
||||
)
|
||||
query_param = QueryParam(mode=mode)
|
||||
|
||||
base_dir='../datasets/questions'
|
||||
base_dir = "../datasets/questions"
|
||||
queries = extract_queries(f"{base_dir}/{cls}_questions.txt")
|
||||
run_queries_and_save_to_json(queries, rag, query_param, f"{base_dir}/result.json", f"{base_dir}/errors.json")
|
||||
run_queries_and_save_to_json(
|
||||
queries, rag, query_param, f"{base_dir}/result.json", f"{base_dir}/errors.json"
|
||||
)
|
||||
|
||||
@@ -1,12 +1,14 @@
|
||||
openai
|
||||
tiktoken
|
||||
networkx
|
||||
accelerate
|
||||
aioboto3
|
||||
graspologic
|
||||
nano-vectordb
|
||||
hnswlib
|
||||
xxhash
|
||||
tenacity
|
||||
transformers
|
||||
torch
|
||||
nano-vectordb
|
||||
networkx
|
||||
ollama
|
||||
accelerate
|
||||
openai
|
||||
tenacity
|
||||
tiktoken
|
||||
torch
|
||||
transformers
|
||||
xxhash
|
||||
pyvis
|
||||
Reference in New Issue
Block a user